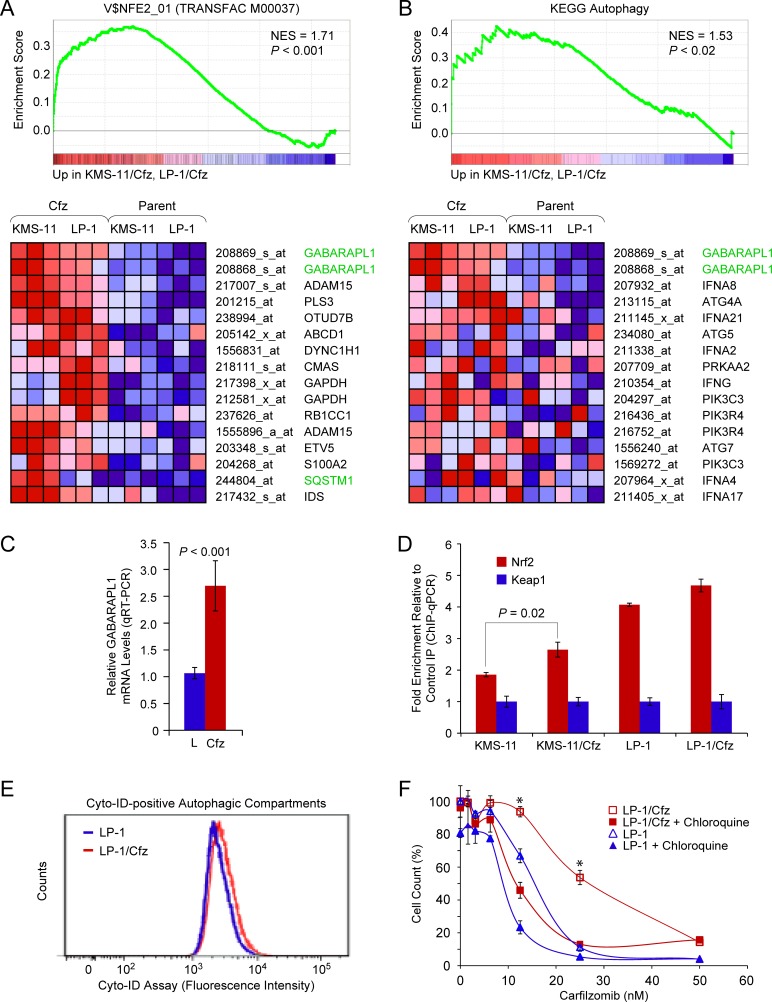
Figure 2. The autophagy-related gene GABARAPL1 is an Nrf2 binding target upregulated in LP-1/Cfz and KMS-11/Cfz cells.
A. GSEA enrichment plot and heat map of the leading edge subset of genes upregulated in both LP-1/Cfz and KMS-11/Cfz cells (triplicate samples) whose promoter regions contain the NF-E2 motif. Gene set: V$NFE2_01 (M1608). The top-ranked probe sets corresponded to GABARAPL1. B. GSEA enrichment plot and heat map of the leading edge subset of autophagy pathway genes upregulated in both LP-1/Cfz and KMS-11/Cfz cells. The top-ranked probe sets corresponded to GABARAPL1. Gene set: KEGG Regulation of autophagy (M6382; KEGG Pathway hsa04140). qRT-PCR analysis was performed to validate the differential expression of GABARAPL1 mRNA in LP-1/Cfz (Cfz) versus parental LP-1 (L) cells (mean values of three qRT-PCR experiments). See Table S1A for expression changes determined from the microarray data. D. Increased binding of Nrf2 to the GABARAPL1 promoter region indicated in Figure S5 in LP-1/Cfz and KMS-11/Cfz cells as determined by ChIP-qPCR. E. Fluorescence histograms of LP-1/Cfz and parental LP-1 cells stained with the Cyto-ID autophagy detection reagent. F. Cells were treated with the indicated concentrations of carfilzomib for 72 hours in the absence or presence of chloroquine (10 μM) and cell viability was determined by alamarBlue assay. *, P < 0.001 vs carfilzomib alone (n = 3).