Figure 1. Diverged DNA-binding “fingerprint” patterns for primate-specific ZSCAN5 proteins.
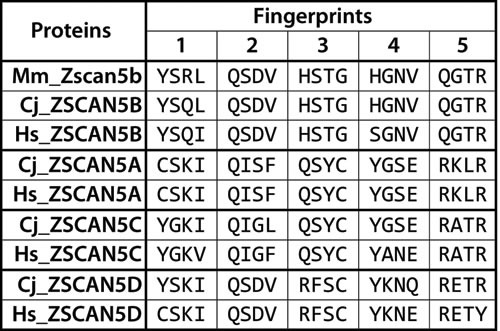
Amino acid residues corresponding to DNA-binding positions (1-, 2, 3, and 6 relative to the alpha helix) in each of the five zinc fingers in ZSCAN5 proteins predicted from mouse (Mm), human (Hs), and marmoset (Cj) genomes are shown aligned in the C- > N terminal order of the zinc fingers in each gene. Mouse and other mammals contain a single gene, Zscan5b, which was duplicated in early primate history to generate three new gene copies: ZSCAN5A, ZSCAN5C, and ZSCAN5D (Mm_Zscan5b: ENSMUSG00000058028, Cj_ZSCAN5B: ENSCJAG00000020279, Hs_ZSCAN5B: ENSG00000197213, Cj_ZSCAN5A: ENSCJAG00000037918, Hs_ZSCAN5A: ENSG00000131848, Cj_ZSCAN5C: ENSCJAG00000020275, Hs_ZSCAN5C: ENSG00000204532, Cj_ZSCAN5D: ENSCJAG00000014787, Hs_ZSCAN5D: ENSG00000267908). The fingerprints of the novel duplicates have diverged relative to the ancestral gene copy, but once generated, these patterns have been conserved in primate species.
