Figure 5. ZSCAN5 binding displays a protein- and cell type-specific preference for tDNAs and other Pol III-related loci enriched in G/C rich motifs.
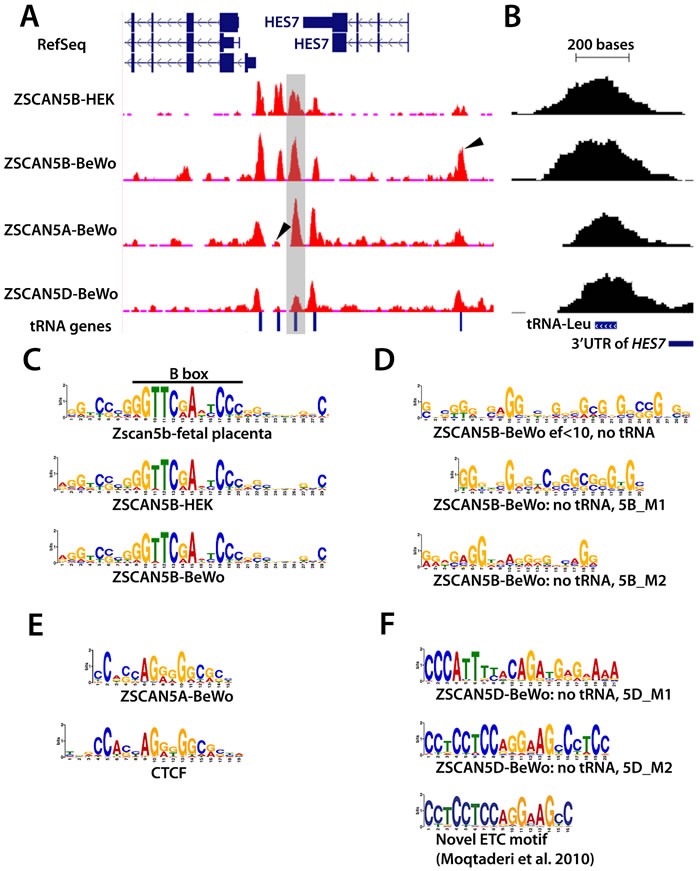
A. Distribution of ChIP-seq reads in the HES7 region is displayed in a snapshot from the UCSC browser. ChIP-seq read pileups are shown in red, with the distribution of background reads from genomic input displayed in brown. Peaks from ZSCAN5B in chromatin from two cell lines, HEK-293 and BeWo, shown together with peak profiles from ZSCAN5A and ZSCAN5D ChIP-seq in BeWo chromatin, reveal strong enrichment over tRNA genes that are clustered in the region. Arrows in peak tracks highlight some of the peaks that are differentially enriched by ZSCAN5B ChIP in the two cell types, or in ChIP with ZSCAN5B compared to ZSCAN5A or ZSCAN5D. The tRNA-Leucine (tRNA-L) gene shown in close-up in panel B is highlighted in grey. B. A close-up view of the tDNA highlighted in grey in panel A, illustrating that the ChIP-seq peak summits are centered over the tDNAs. C. Motifs detected as most highly enriched and central in ChIP-seq peaks for mouse Zscan5b and human ZSCAN5B in HEK-293 or BeWo cells include the TFIIIC-binding B-box, which is present and highly conserved in all expressed tDNAs, and strikingly similar tracts of G/C-rich surrounding DNA. D. A motif search conducted after removing tDNA sequences from the ZSCAN5B BeWo peak set also revealed G/C-rich sequences, including an extended motif comprising two shorter motifs (5B_M1, 5B_M2) as predicted ZSCAN5B binding motifs. E. A G/C rich central motif was also detected in analysis of ZSCAN5A ChIP peaks; a portion of this motif bears striking resemblance to the known motif for transcription factor and insulator protein, CTCF. F. A distinct set of motifs were detected in high-scoring ZSCAN5D peaks, including a G/C rich motif, 5D_M2, which bears striking similarity to a novel ETC motif detected by Moqtaderi and colleagues [19].
