Figure 1. Four principal roles of autophagy in immunity.
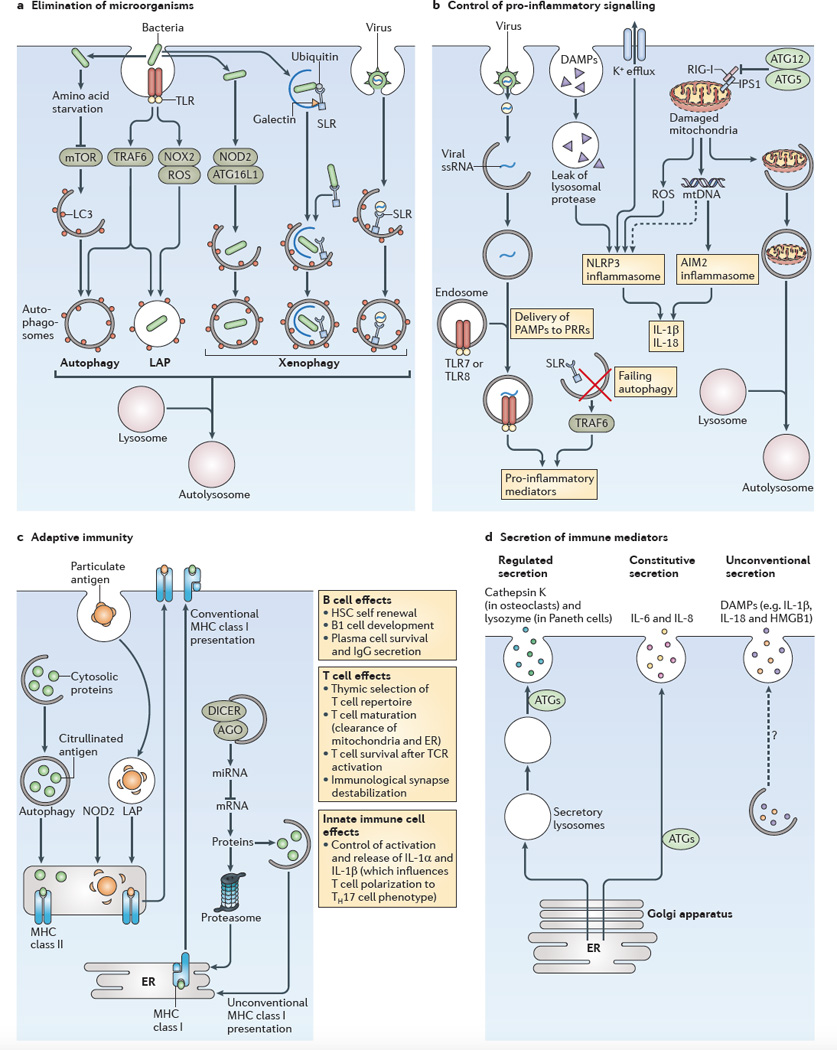
a | The role of autophagy in the elimination of microorganisms is shown. An incoming microorganism can induce autophagy by competing for nutrients or by stimulating innate immune receptors, such as Toll-like receptors (TLRs). When the microorganism is taken up by phagocytosis and remains in an intact vacuole, an autophagic process termed LC3-associated phagocytosis (LAP) can promote the maturation of autophagosomes into autolysosomes. Xenophagy of pathogens that enter the cytosol can be initiated by sequestosome 1-like receptors (SLRs) or other mechanisms, including nucleotide-binding oligomerization domain-containing protein 2 (NOD2)–autophagy-related protein 16-like 1 (ATG16L1) interactions. b Several examples of the role of autophagy in the control of pro-inflammatory signalling (see also FIG. 3) are shown. Failure to remove SLRs by autophagy can increase the levels of these receptors and the levels of pro-inflammatory signalling. Autophagy can deliver cytoplasmic pathogen-associated molecular patterns (PAMPs) to endocytic TLRs and can stimulate their activity. NOD-like receptors (NLRs; such as NLRP3 (NOD-, LRR- and pyrin domain-containing protein 3)) and RIG-I-like receptors (RLRs; such as absent in melanoma 2 (AIM2)) show complex positive and negative co-regulation with autophagy: the ATG5–ATG12 complex inhibits retinoic acid-inducible gene I (RIG-I) signalling, and autophagy limits inflammasome activation by removing damaged mitochondria, which then release the inflammasome activators reactive oxygen species (ROS) and mitochondrial DNA. c The role of autophagy in adaptive immunity is shown. Autophagy can increase the MHC class II presentation of cytoplasmic antigens, including self or viral antigens, as well as promoting the citrullination of antigens. LAP can enhance the processing of particulate antigens for MHC class II presentation. NOD2 enhances autophagic antigen presentation. Autophagy may directly or indirectly affect MHC class I presentation by competing with the proteasome for substrates, by influencing the peptidome pools through the control of levels of components of microRNA (miRNA) machinery (for example, argonaute (AGO) and DICER), or by supporting unconventional MHC class I presentation. In addition, autophagy affects the self-renewal of haematopoietic stem cells (HSCs), B1 cell development, plasma cell survival and IgG secretion. Autophagy affects T cell survival following T cell receptor (TCR) activation, and it destabilizes the immunological synapse. It also controls innate immune cell (such as macrophage) signalling through the release of interleukin-1α _(IL-1α)_ _and IL-1β, which influence the polarization of T cells into T helper 17 (TH17) cells. Autophagy also affects naive T cell repertoire selection in the thymus and the survival and function of maturing T cells by removing the mitochondria and endoplasmic reticulum (ER), thus ensuring calcium homeostasis. d The role of autophagy in the secretion of immune mediators is shown. Autophagy affects the quality of regulated secretion from pre-stored granules. Autophagy affects the quality and the quantity of the output of the constitutive secretory pathway (which is the conventional pathway of protein secretion via the ER, the Golgi apparatus and the plasma membrane). Autophagy supports a form of unconventional secretion that captures cytoplasmic proteins for extracellular release. Note that secretory protein cargo in the regulated and constitutive secretory pathways contains conventional leader peptides for co-translational import into the ER lumen, whereas protein cargo that enters the unconventional secretory pathway lacks leader peptides and does not enter the ER. Dashed arrow indicates that this pathway remains to be defined. DAMPs, damage-associated molecular patterns; HMGB1, high-mobility group box 1 protein; IPS1, IFNβ _promoter stimulator protein 1; mtDNA, mitochondrial DNA; mTOR, mammalian target of rapamycin; NOX2, NADPH oxidase 2; PRRs, pattern recognition receptors; ssRNA, single-stranded RNA; TRAF6, TNF receptor-associated factor 6.
