Figure 2. Autophagy-mediated clearance of intracellular pathogens.
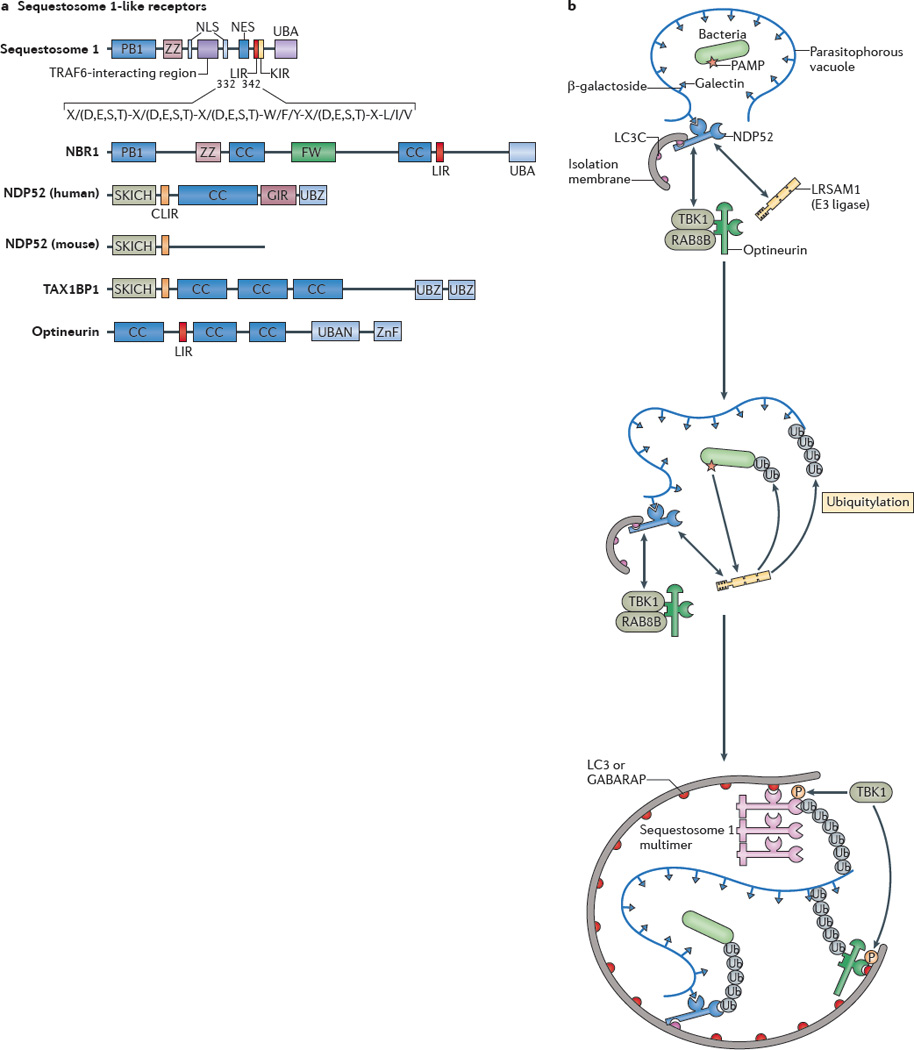
a| Protein domains of sequestosome 1-like receptors (SLRs) are shown. The LC3-interacting region (LIR) motif of an SLR binds to autophagy-related LC3 proteins through its consensus sequence at amino acids 332–342 in sequestosome 1 (also known as p62). The conserved residues are shown. X/(D,E,S,T) indicates that any amino acid (X) is allowed but that acidic (D,E) or phosphorylatable amino acids (S,T) are often present (usually at least one or more within the entire consensus sequence). The core LIR motif residues are aromatic pocket-filling W (or F or Y) residues and aliphatic pocket-filling L (or I or V). They form an intermolecular parallel β-_sheet with LC3 proteins or γ-aminobutyric acid receptor-associated proteins (GABARAPs). The CLIR motif, which is a LIR motif that is specific for LC3C, lacks the aromatic residue found in the LIR motif and, instead, uses hydrophobic contacts provided by additional aliphatic residues located between the W and L position anchors to stabilize interactions with LC3C. All human SLRs also contain a ubiquitin-binding domain (UBD): UBA (as found in sequestosome 1 and NBR1) is a three-helix bundle UBD that has affinity for monoubiquitin and K63 ubiquitin linkages; UBAN (as found in optineurin) is a parallel coiled-coiled dimer UBD that has specificity for linear ubiquitin chains; and UBZ (as found in nuclear dot protein 52 (NDP52)) is a zinc finger ββα-fold UBD that binds to monoubiquitin and polyubiquitin. b A model of cooperative action between different SLRs and E3 ligases in bacterial targeting for xenophagy is shown. The schematic shows a parasitophorous vacuole with glycosylated molecules (in this case β-galactosides) facing the lumen of the vacuole that contains a bacterium and that is experiencing membrane damage. This membrane tear exposes β-galactosides to galectins (for example, galectin 8) which in turn bind to the galectin-interacting region (GIR) motif of NDP52. NDP52 also directly interacts with the E3 ligase LRSAM1 and indirectly with the serine/threonine protein kinase TANK-binding kinase 1 (TBK1), which interacts with optineurin. The CLIR motif of NDP52 binds to LC3C, which is a proposed initiator in the LC3 and GABARAP cascade during bacterial xenophagy. LRSAM1 or other E3 ubiquitin ligases polymerize ubiquitin at molecular targets that are yet to be identified. The hypothetical model includes the putative recognition of bacterial pathogen-associated molecular patterns (PAMPs) by LRSAM1 through its leucine-rich repeat domain. Ubiquitin tags are recognized by UBDs of NDP52, optineurin and sequestosome 1. The LIR motif of optineurin is phosphorylated by TBK1 and this improves LC3 and GABARAP binding. The UBA of sequestosome 1 is also phosphorylated by TBK1 and this improves ubiquitin chain binding. As a consequence, the autophagic isolation membrane initiates at the appropriate location and grows to capture the bacterium and to eliminate it through autophagy. CC, coiled-coil domain; FW, four W domain (also known as the NBR1 domain); KIR, KEAP1 interacting region; NES, nuclear export signal; NLS, nuclear localization signal; P, phosphorylation; PB1, protein-binding domain 1; SKICH, skeletal muscle and kidney enriched inositol phosphatase carboxyl homology domain; TAX1BP1, TAX1-binding protein 1; Ub, ubiquitylation; ZnF, zinc finger domain; ZZ, ZZ-type ZnF domain.
