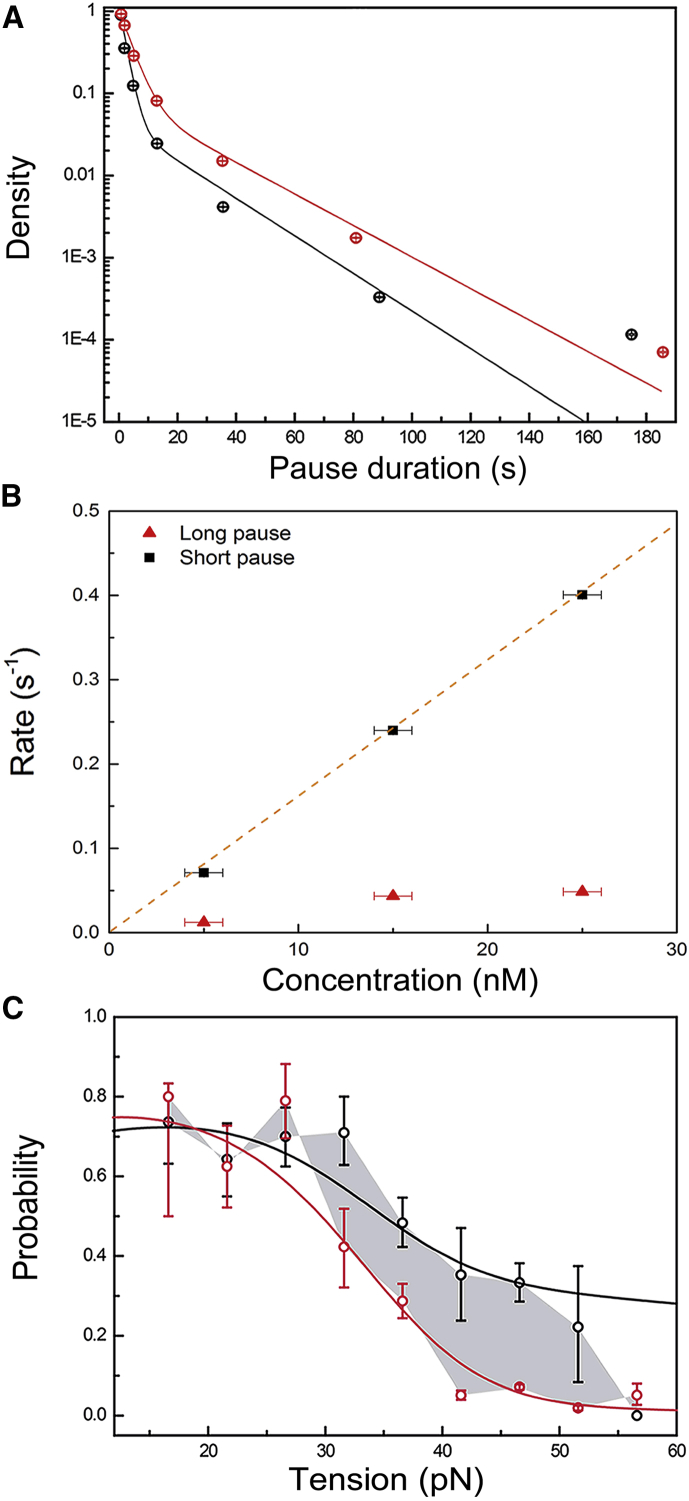
Figure 3.
Distribution of pause durations. (A) Black, high concentration (25 nM, 1535 pauses, all tensions); red, low concentration (15 nM, 1430 pauses, all tensions); solid lines, double-exponential fits. For clarity, the data and fits have been normalized to 1 at 0.4 s. The error bars represent the 95% confidence interval of 10,000 bootstrapped data sets. (B) Binding rates for three concentrations (1000 bootstraps). The binding rate is the inverse of the characteristic pause duration. Black square, short pause; red triangle, long pause; 25 nM (1535 pauses, all tensions); 15 nM (1430 pauses, all tensions); 5 nM (313 pauses, 30 pN). The error bars in x reflect the error in concentration we expect due to absorption of protein on the walls of our flow chamber. The y error bars are smaller than the dots indicating the data. The orange dashed line is a linear fit of the short pause rates and is drawn through zero to guide the eye. (C) The probability of binding in the pol active site after a pause preceded by polymerization (black) or exonucleolysis (red) activity (mean ± SD, data for all concentrations and tensions are combined). The gray area marks the difference between the probabilities. The solid lines are not a direct fit to these data, but are calculated using the rates fitted to the complete time statistics of pauses (see Supporting Material).