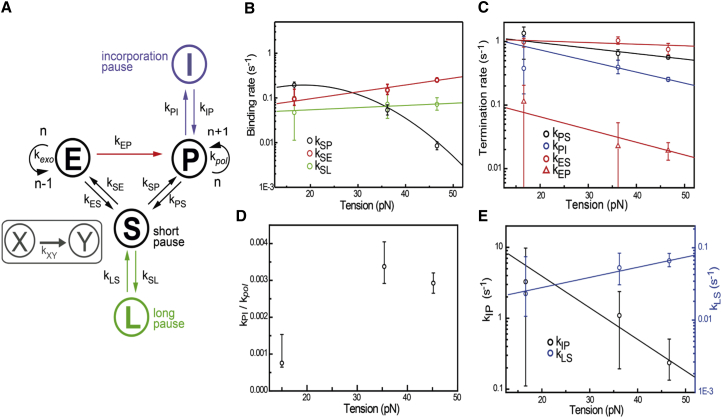
Figure 4.
Kinetic model of T7 DNAp fitted to the experimental data. (A) Scheme of the kinetic model, with rates indicated. The original model (based on biochemical data (15)) is indicated in black. On the basis of our experimental results, three modifications were made. Green, long-pause state; blue, incorporation pause; red, direct switching occurring only from the exo to the pol active site. (B–E) Results of maximum-likelihood fitting of the model in (A) to the experimental data. Here we choose to bin our data in three forces (10, 35, and 45 pN) to minimize the error bars, allowing us to constrain our fits optimally. Using more bins would increase the error bars while not providing much additional information about the general trend of the data. Sensitivity in the parameters is estimated by bootstrapping the data (300 times), and the 95% confidence interval is shown. Except for kSP, all rates are fitted with . kSP is fit to a second-order force dependent rate. (B) Binding rates (kSP, kSE, kSL) out of solution as function of tension. (C) Off-rates of polymerization and exonucleolysis activities (kPS, kPI, kES), and the direct-switching rate from exo to pol (kEP) as a function of tension. (D) Ratio of the rate into the incorporation pause (kIP) over the polymerization rate (kPol) as a function of tension. (E) Recovery rates from incorporation pauses (kPI) and long pauses (kLS) as a function of tension.