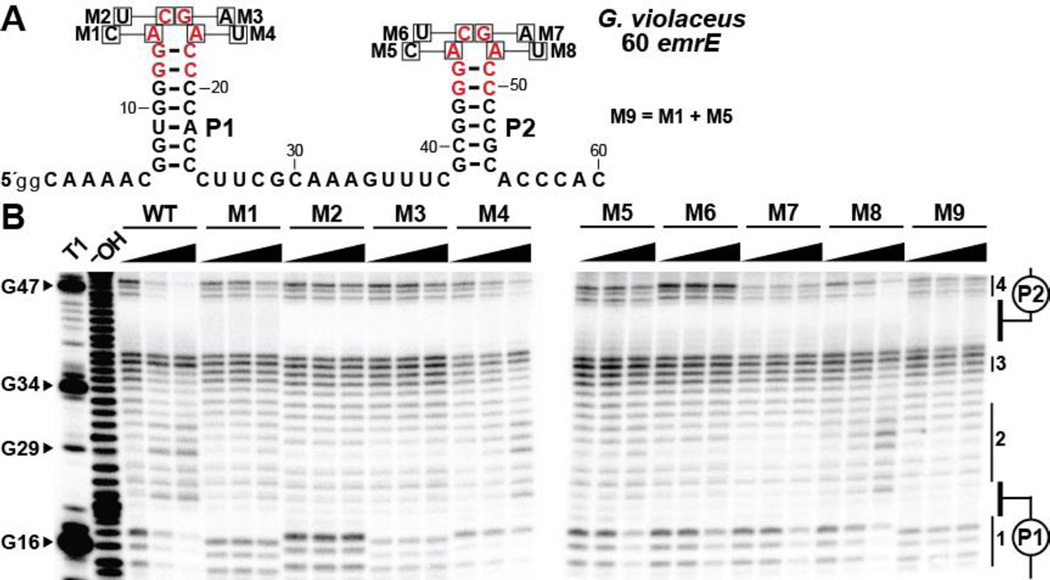
Figure 3.
Mutations of conserved loop nucleotides affect structural modulation of both loops. (A) Sequence and secondary structure of the 60 emrE RNA from G. violaceus. Red letters denote highly conserved (>97%) nucleotide positions of the consensus sequence. Single nucleotide positions altered to create each RNA are identified with boxes, annotated with the mutant nucleotide identity, and labeled M1 through M8. M9 contains mutations at both positions specified by M1 and M5. (B) PAGE analysis of in-line probing assays with WT and mutated sequences of the 60 emrE RNA in the presence of 0, 0.1, and 1 mM guanidine. The cropped gel image depicts the regions spanning from loop 1 (nucleotides 14–17) to loop 2 (nucleotides 45–48). Additional annotations are as described in the legend to Figure 2B.