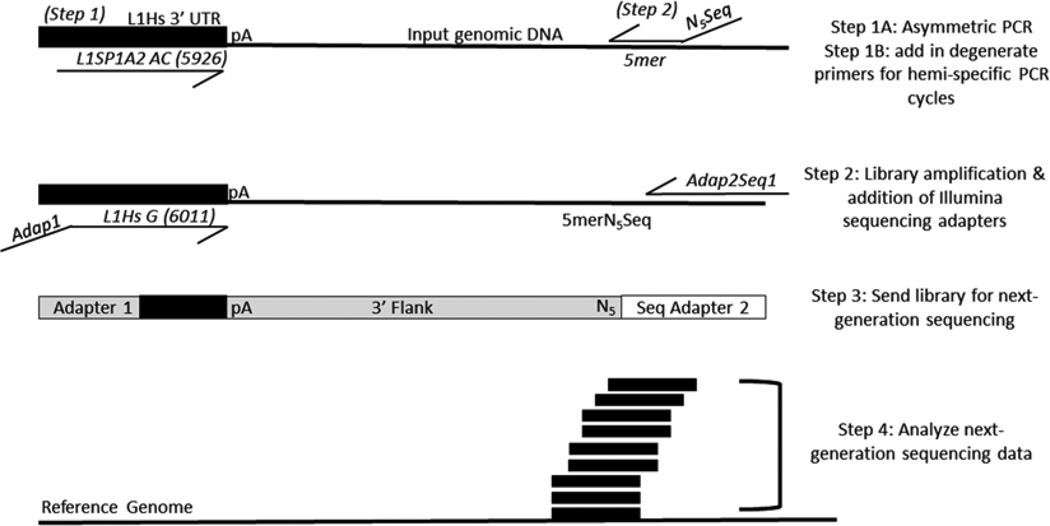
Fig. 1.
Scheme of L1-seq. The first PCR consists of five cycles which enrich for sequences containing human-specific L1 sequences via primer extension with the L1Sp1A2 primer using diagnostic nucleotides for the human specific subfamily of L1. After enrichment for human-specific L1 flanks in the first five cycles, each reaction has one degenerate primer added. There are eight degenerate primers, each with a specified 5-mer at the 3′ end preceded by five degenerate bases (NNNNN) and a sequencing primer used for the Illumina platform. Eight different reactions are performed, each with a unique 5mer. The second PCR enriches for human-specific L1 3′ flanks by utilizing another diagnostic nucleotide in the 3′ UTR of L1 and adds the necessary adapter sequences via primer overhangs. The products of this reaction are mixed in equimolar ratios before sequencing on the Illumina 2500 platform. Following sequencing and initial processing, the reads are aligned to the human genome (hg19) and L1 insertions are identified for validation