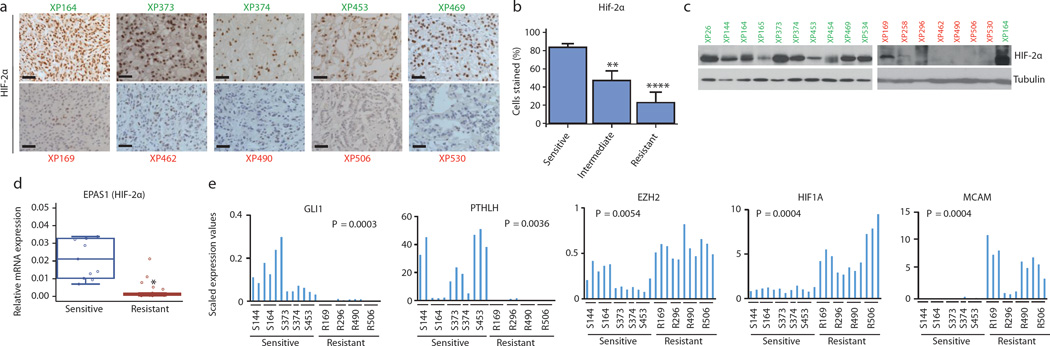
Fig. 3. Sensitive and resistant tumors can be distinguished by HIF-2α levels and gene expression signature.
a, HIF-2α expression by immunohistochemistry (IHC) in sensitive (green) and resistant (red) tumors. Scale bars = 50 µM. (Images representative of quantitative data shown in 3b.) b, Quantification of HIF-2α-positive cells as determined by IHC in sensitive, intermediate, and resistant tumors from all 22 tumorgraft lines (Sensitive: n = 10; Intermediate: n = 5; Resistant: n = 7). c, Western blot analysis of sensitive (green) and resistant (red) tumorgraft lines. XP164 lysate loaded twice as a reference for comparison between the two membranes. d, qRT-PCR of EPAS1 (HIF-2α) expression in sensitive (n = 11) versus resistant (n = 26) vehicle-treated tumorgrafts. e, Candidate genes from RNAseq analysis differentially expressed in sensitive and resistant tumors. b: An ANOVA test was used to determine if sensitive tumors were different from intermediate or resistant. Bar chart depicts the mean with the error bar representing s.e.m. d, e: Tests completed using a mixed model analysis with compound symmetrical covariance structure for mice in the same tumorgraft line. RNAseq values were log2-transformed for analysis; Raw values depicted in all graphs. Bar charts depict individual RNA-seq values, while all boxplots have median centre values. **, p < 0.01; and ****, p < 0.0001. See Supplementary Fig. 1 for gel source data.