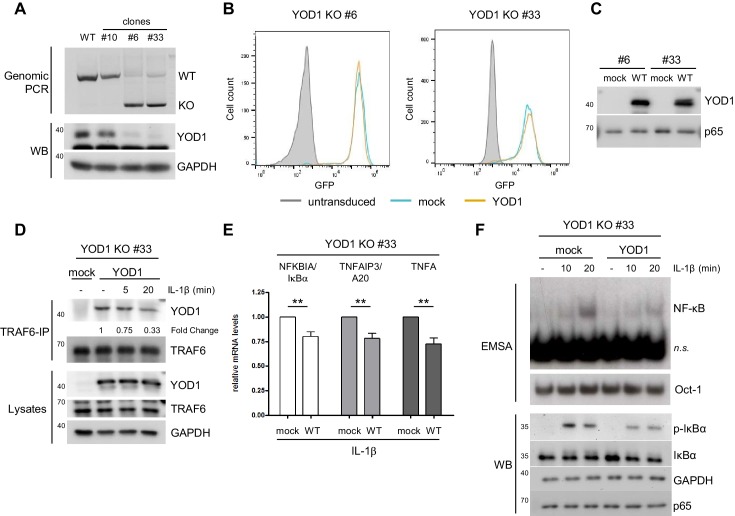
Figure 5. Reconstitution of YOD1-deficient HeLa cells impairs IL-1β-induced NF-κB signaling.
(A) Validation of YOD1 KO HeLa cell clones. YOD1 genomic DNA and protein levels in parental HeLa cells and in cell clones generated by CRISPR/Cas9 gene editing were checked by PCR and Western Blot. (B) YOD1 deficient HeLa clones #6 and #33 are efficiently transduced with empty vector (mock) and YOD1 WT. Cells were transduced and homogenous populations of GFP expressing cells were sorted by FACS. FACS of GFP expression after sorting is shown. (C) Reconstitution of YOD1-deficient cell clones #6 and #33 with YOD1 WT. YOD1-deficient HeLa cells were transduced with YOD1 WT or mock constructs and YOD1 expression was analyzed by Western Blot. (D) YOD1/TRAF6 interaction in reconstituted YOD1-deficient HeLa clone #33 is decreasing upon IL-1R engagement. Cells were stimulated with IL-1β for the indicated time points. Anti-TRAF6 IPs were conducted and interaction of YOD1 was analyzed by Western Blot. Quantification of YOD1 bound to TRAF6 is shown. Numbers indicate the fold change after IL-1β stimulation (unstimulated set to 1). (E) Reconstitution of YOD1-deficient HeLa clone #33 with YOD1 WT diminishes NF-κB target gene expression. Cells were stimulated with IL-1β for 40 min. RNA was isolated and transcripts were analyzed by qRT-PCR as indicated. Bars show mean and SEM of seven independent experiments. Significance was evaluated using Student’s t-test (*p<0,05; **p<0,01; ***p<0001; ns = not significant). (F) YOD1 re-expression in YOD1-deficient HeLa clone #33 diminishes NF-κB activation and IκBα phosphorylation and degradation. Cells were stimulated with IL-1β for the indicated time points and NF-κB DNA binding was assessed by EMSA (n.s. = non-specific band). Oct-1 EMSA served as loading control. IκBα phosphorylation and degradation was analyzed by Western Blot.