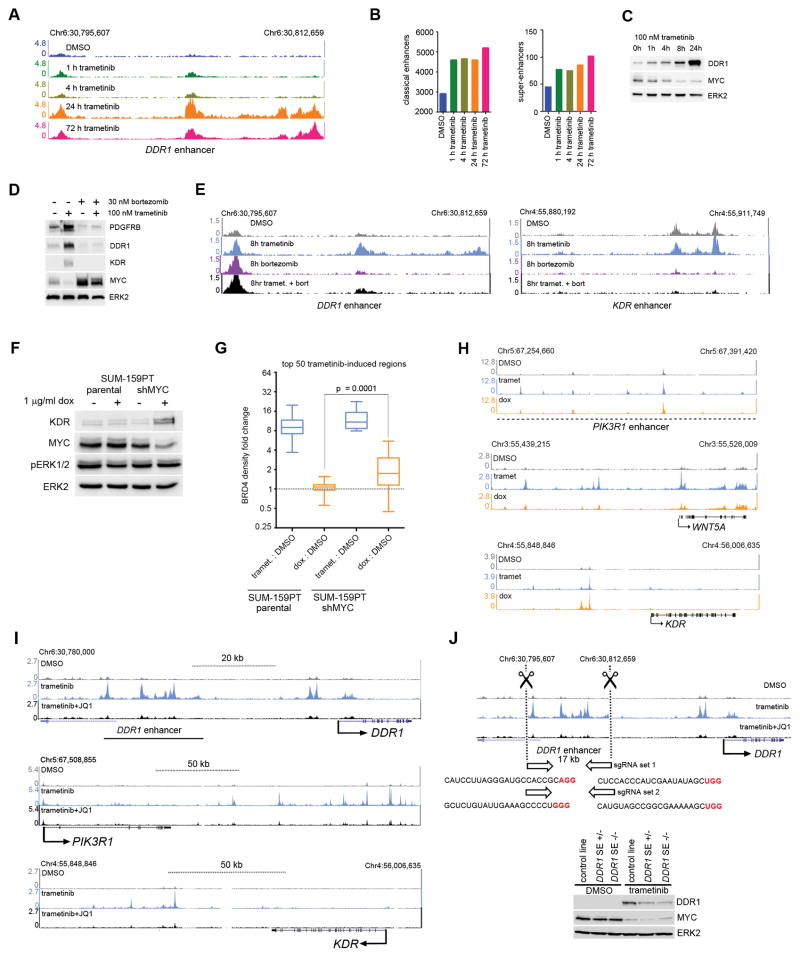
Figure 4.
Proteasome or BET bromodomain inhibition attenuates trametinib-induced enhancers at kinase loci. (A) Time course of BRD4 density induced by 100 nM trametinib treatment at the DDR1 enhancer. (B) Classical enhancer (left) or super-enhancer (right) quantification by BRD4 density over 100 nM trametinib time course. (C) Time course of Myc protein levels following trametinib treatment showing anticorrelation of DDR1 protein induction and BRD4 density (A) or enhancer induction (B). (D) Western blot showing Myc stabilization and loss of PDGFRB, DDR1, and KDR upregulation with 8 h co-treatment of 100 nM trametinib and 30 nM bortezomib. (E) Loss of trametinib-induced DDR1 (left) and KDR (right) BRD4 enhancer density upon co-treatment with 30 nM bortezomib. (F) Upregulation of adaptive response RTK KDR upon doxycycline induction of Myc shRNA in stable SUM-159PT cells. (G) BRD4 density change at the highest ranking 50 trametinib-induced regions in response to 48 h 100 nM trametinib or 1 μg/ml doxycycline induction of Myc shRNA. (H) BRD4 density induction following 48 h 100 nM trametinib or 1 μg/ml doxycycline induction of Myc shRNA at PIK3R1, WNT5A or KDR1 adaptive response loci. (I) BRD4 ChIPseq density tracks depicting enhancer formation following 24 h 100 nM trametinib and enhancer blockade following co-treatment with 300 nM JQ1 at the DDR1, PIK3R1, and KDR SUM-159PT adaptive response genes. (J) Top: CRISPR/Cas9 deletion of the SUM-159PT DDR1 trametinib-induced enhancer. Bottom: Attenuation of DDR1 protein induction following 24 h 100 nM trametinib in stable SUM-159PT cell lines either heterozygous or homozygous for the enhancer deletion.