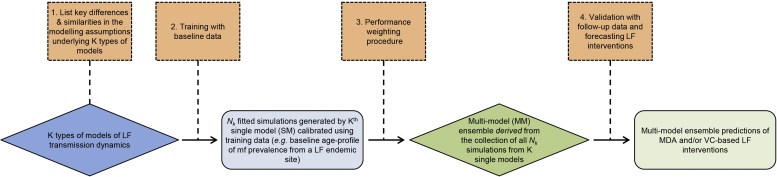
Fig. 1.
Overview of the methodology used for developing the lymphatic filariasis (LF) multi-model ensemble. All K (=3) single LF models were 1) compared prior to inclusion and 2) trained with baseline data on LF infection (Table 2A) to produce a collection of Nk simulations for each of the three LF model types. Each constituent model was 3) assigned a weight reflecting its relative performance in reproducing the baseline age-mf prevalence data in each site. The weights were used to construct the LF multi-model ensemble. To validate the multi-model ensemble and forecast the effects of LF interventions, 4) simulations were generated by the multi-model ensemble and compared with mf age-prevalence data obtained during the intervention period in each site (Table 2B). The four processes outlined are represented by orange boxes. The different types of models (SM and MM) are represented as blue and green diamonds, respectively, and the corresponding simulation outcomes are represented as rectangles of the same color.