FIGURE 3.
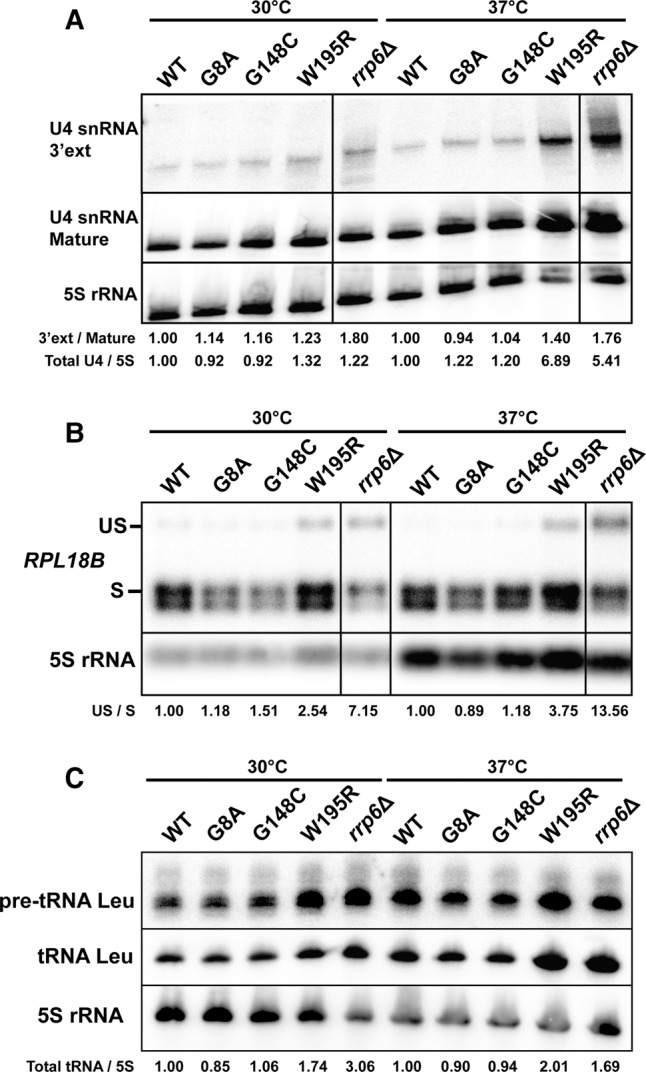
snRNA processing and degradation defects of Rrp40p mutants. (A) Analysis of U4 snRNA processing. Membranes were hybridized with an oligonucleotide probe downstream from the U4 snRNA or to an oligonucleotide hybridizing to the mature U4 snRNA. Lines drawn on the Northern blots indicate that lanes from these blots were removed from the picture. However, all the lanes shown for each panel originate from the same membranes and hybridizations. The 5S rRNA was used as a loading control. The values shown indicate the ratio of U4 3′-extended species to mature species, normalized to that ratio for the respective WT sample at each temperature. Also provided are the total U4 signals normalized to the 5S rRNA signals, which are in turn normalized to that ratio for the respective WT sample at each temperature. (B) Analysis of RPL18B spliced and unspliced RNA levels. The 5S rRNA was used as a loading control. Quantitation of unspliced to spliced (US/S) RPL18B was performed similarly to A. (C) Analysis of Leu pre-tRNA and tRNA levels. The 5S rRNA was used as a loading control. Quantitation of total tRNA signals was performed similarly to A and B.
