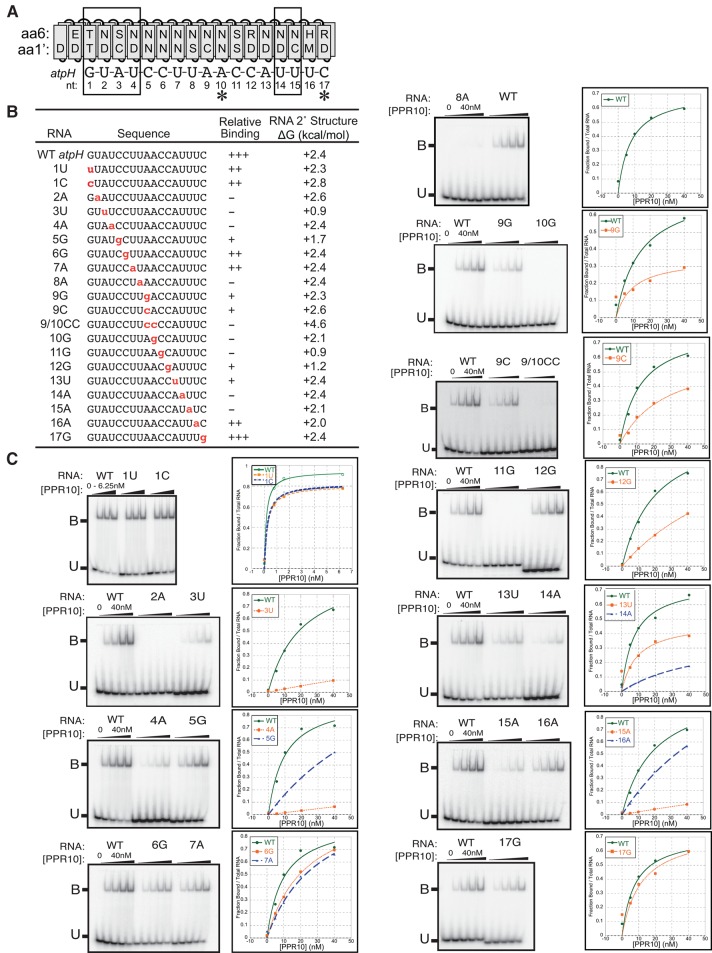
FIGURE 3.
Effects of single-nucleotide changes in the atpH site on PPR10 binding affinity. (A) Alignment between PPR10 and the atpH binding site. Boxes mark the modular PPR–nucleotide contacts detected in the PPR10–psaJ crystal structure (Yin et al. 2013) and asterisks mark nucleotides that make nonmodular protein contacts in that structure. (B) RNAs used for gel mobility shift assays. Nucleotide substitutions are marked in red. Relative binding affinities were approximated based on the data shown in panel C; mutant RNAs were placed into relative affinity bins by comparing their binding behavior to that of the wild-type RNA when assayed with the identical protein dilutions on the same gel. The thermodynamic stability of RNA secondary structure predicted for each sequence (Mfold prediction [Zuker 2003], 37°C, default parameters) is shown to the right. (C) Gel mobility shift assays. PPR10 was used at the concentrations indicated in the graphs. All assays used the same preparation of PPR10 except the assays with 1U and 1C RNAs. Each assay was repeated either two or three times, with similar results. A representative assay is shown in each case.