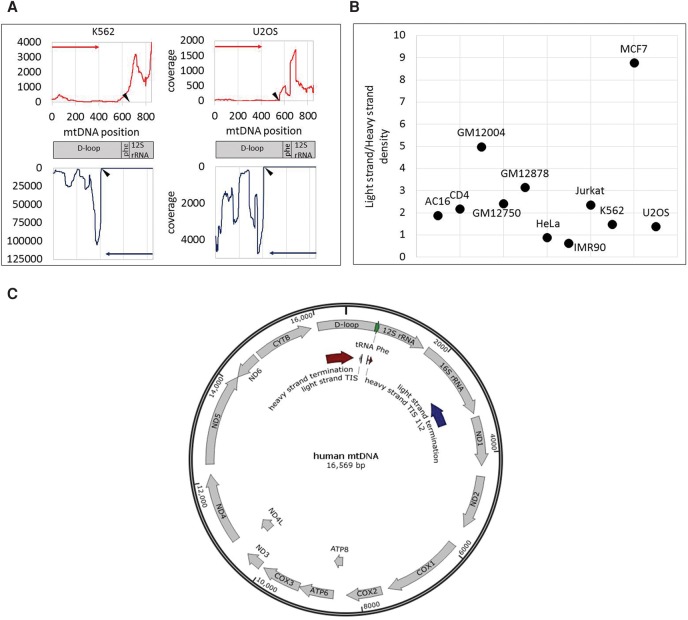
Figure 2.
Accurate identification of human mtDNA TIS. (A) Sequencing read coverage around the mtDNA TIS in two cell lines. (Top) Sequencing read coverage pattern of the mtDNA heavy strand (red); (bottom) sequencing read coverage of the light strand (blue). Putative TIS is designated by black triangle. (y-axis) Sequencing read coverage; (x-axis) mtDNA position. (Left) PRO-seq experiment of the K562 cell line. (Right) GRO-seq experiment from the U2OS cell line. (B) Ratio of sequencing coverage between the light and heavy strands. (y-axis) Ratio of read density between the light and heavy strands. Black dots correspond to the calculated ratios for each tested cell line (indicated near the dots). (C) Summary of mtDNA transcription pattern: PRO-seq and GRO-seq experiments in 11 human cell types: (TIS) Light-strand TIS was identified in all tested human cell types (N = 11) in positions 407–410. In most of the tested cells (seven of 11), the major heavy-strand TIS was mapped in positions 560–562 (TIS 1). In the remaining cell types (four), the major heavy-strand TIS was located in positions 634–689 (TIS 2). (Termination sites) Light-strand transcription termination was identified within the 16S rRNA gene, in the region encompassing positions 2612–3252 (dark blue arrow). Heavy-strand transcription termination was identified within the D-Loop, between positions 16,076 and 195 (dark red arrow).