Fig. 2.
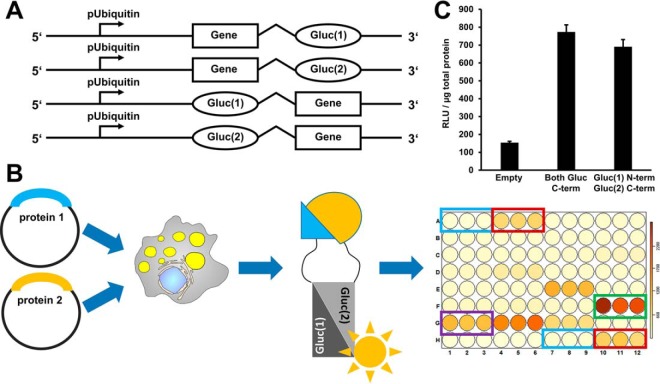
A protein-protein interaction assay based on luciferase fragment complementation. A, On the basis of a published luciferase fragment complementation assay, we constructed Gateway recombination cloning vectors suitable for the expression of luciferase fragment fusion proteins in Drosophila cells. These fusion proteins carry at the N- or C terminus either the N-terminal half of the Gaussia princeps luciferase protein (Gluc(1)) or the C-terminal half of the luciferase protein (Gluc(2)). B, In order to test for protein interactions, two plasmids encoding the potential interaction partners fused to Gluc(1) or Gluc(2), respectively, are coexpressed in cells. If the proteins interact, the luciferase fragments can assemble into an enzymatically active enzyme. Luciferase activity is measured in a 96-well plate assay format using an injector equipped luminometer plate-reader. Our standard assay plate design covered every condition/plasmid combination in triplicate. We included untransfected cells as negative controls (blue rectangle), coexpression of a leucine zipper as a positive control and for thresholding purposes (red rectangle), and the interaction between the Jabba and CG9186 proteins as a LD specific positive control (green rectangle). Protein combinations resulting in luciferase activity reads exceeding a certain threshold were scored as interaction (e.g. purple rectangle). For the different thresholding levels cf. material and methods. C, We used the dimerization of the GCN4 leucine zipper protein as a positive control. Luciferase complementation readings normalized to total protein content (RLU/μg total protein) were about ten times higher than background levels of untransfected cells. In this case, luciferase fragment positioning did not have a prominent influence on the results.