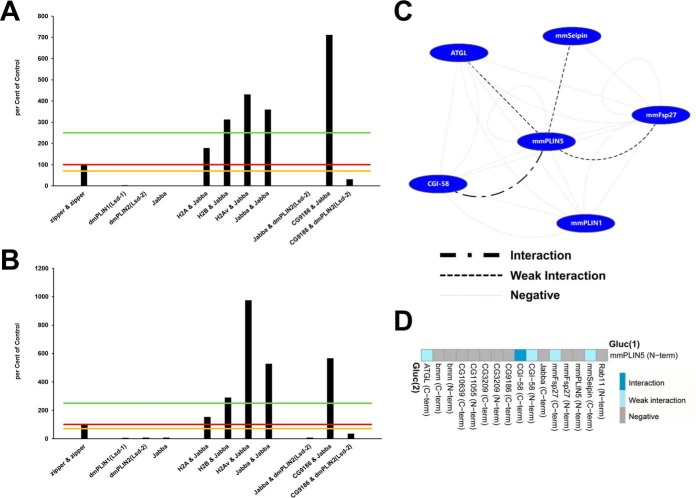
Fig. 3.
The split luciferase complementation assay shows excellent reproducibility and dynamic range. In order to test the performance and reproducibility of the Gaussia princeps split luciferase complementation PPI assay, we transfected cells with the same set of constructs in two biologically independent experiments (A, B): the known leucine zipper interaction pair, single expression plasmids—which thus could not result in a complementation and serve as a negative control—as well as several cotransfections with interacting as well as noninteracting protein pairs. Luciferase reads were normalized to total cellular protein and expressed as percent of the leucine zipper reference protein-protein interaction results. Different significance levels for interaction scoring were introduced: 0 to 70% of zipper-zipper—negative interaction; 70–100% of zipper-zipper—weak interaction; 100–250% of zipper-zipperinteraction; above 250% of zipper-zipper—strong interaction. Significance threshold levels are marked in (A, B) by orange, red, and green lines, respectively. The replicate experiment resulted in concordant scoring of the respective interaction tests. Absolute values, however, can differ between independent experiments (e.g. H2Av&Jabba, or CG9186&Jabba). C, Interactions among mammalian LD-associated proteins. Nodes represent proteins, edges symbolize luciferase complementation test results. A legend for the different edge types is provided. D, Luciferase complementation assay results for the murine PLIN5 protein tagged at the N terminus with the Gluc(1) fragment and different putative interactors. A color-code of the complementation assay results is provided. For protein combinations tested multiple times, the highest luciferase complementation result is visualized.