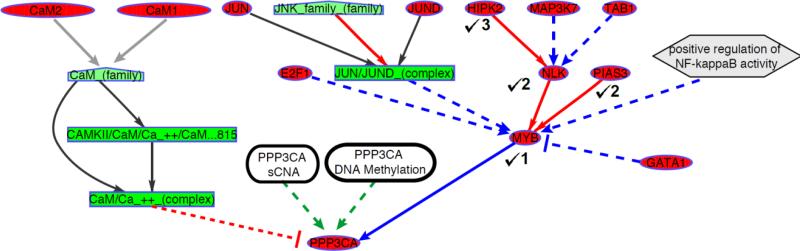
Fig. 1.
An example of a tree (dmax = 3) generated using the regulatory interactions derived from pathway databases for a sample gene PPP3CA. Node types are denoted as Genes (ovals), Protein complexes (rectangles), gene families (pentagon), abstract concepts (hexagon). The edges are colored according to their regulatory function with protein activation (red), transcriptional regulation (blue), component of protein complex (black) and gene family member (grey). The root node's epigenetic and sCNA measurements (rounded rectangles), considered as additional regulatory parents, are connected by green arrows. The nodes chosen as regulators are marked ( ) along with their depths.
) along with their depths.