Fig. 3.
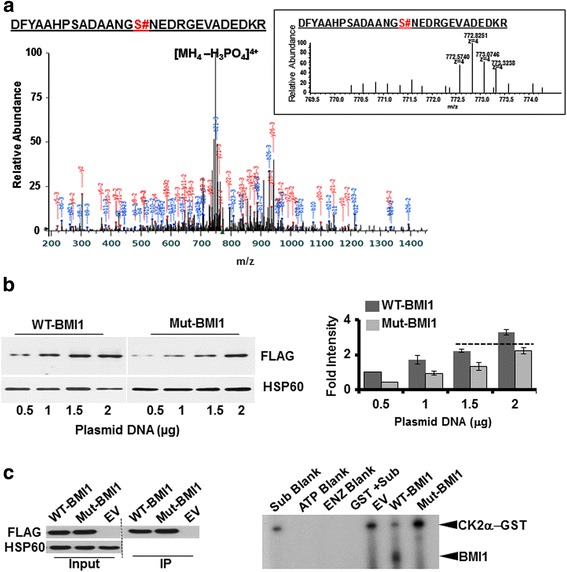
Identification of amino acid residue on BMI1, phosphorylated by CK2α: a Kinase assay and Mass-spectrometric analysis. Purified BMI1 and CK2α were allowed to react in a kinase assay buffer supplemented with nonradioactive ATP. The reaction was terminated after 30 min and the sample was sent for mass spectrometric analysis. The spectrum of the BMI1 phospeptide is presented with the extracted ion chromatogram for the peptide with phosphate shown in the inset (indicated by #). b Relative expression of WT and mutant BMI1: Phospho mutant BMI1 with substitution of Serine at 110 with Alanine (S110A mutant BMI1) was generated using the wild-type BMI1 (WT-BMI1) as a template for site directed mutagenesis. CP20 cells were transfected with increasing DNA concentration (0.5 μg -2 μg) of wild-type or mutant BMI1 and 30 h post transfection samples were harvested for immunoblotting with FLAG antibody. HSP60 is used as a loading control. Right panel shows a representative immunoblot for the expression of the WT and Mut-BMI1. To determine DNA concentration required to achieve comparable expression between WT and Mut-BMI1, a densitometry analysis of the bands was performed using Image J software (NIH, Bethesda, MD), and the graph was generated by plotting fold intensity vs DNA concentration. The broken line indicates equivalent expression level of WT and Mut-BMI1. c Kinase assay with Mutant BMI. CP20 cells were transfected with EV (2 μg), WT (1.5 μg) and Mut-BMI1 (2 μg) and 30 h post transfections cells were harvested for immunoprecipitation with FLAG antibody and agarose beads. A fraction of the agarose beads were processed for immunoblotting to ensure efficient immunoprecipitation and depicted as an immunograph in the left panel. The right panel shows radiograph for kinase assay reaction with the immunoprecipitated beads and in presence/absence of Purified GST-CK2α and radiolabeled ATP
