Figure EV1. Interaction of the Usp27x with BimEL .
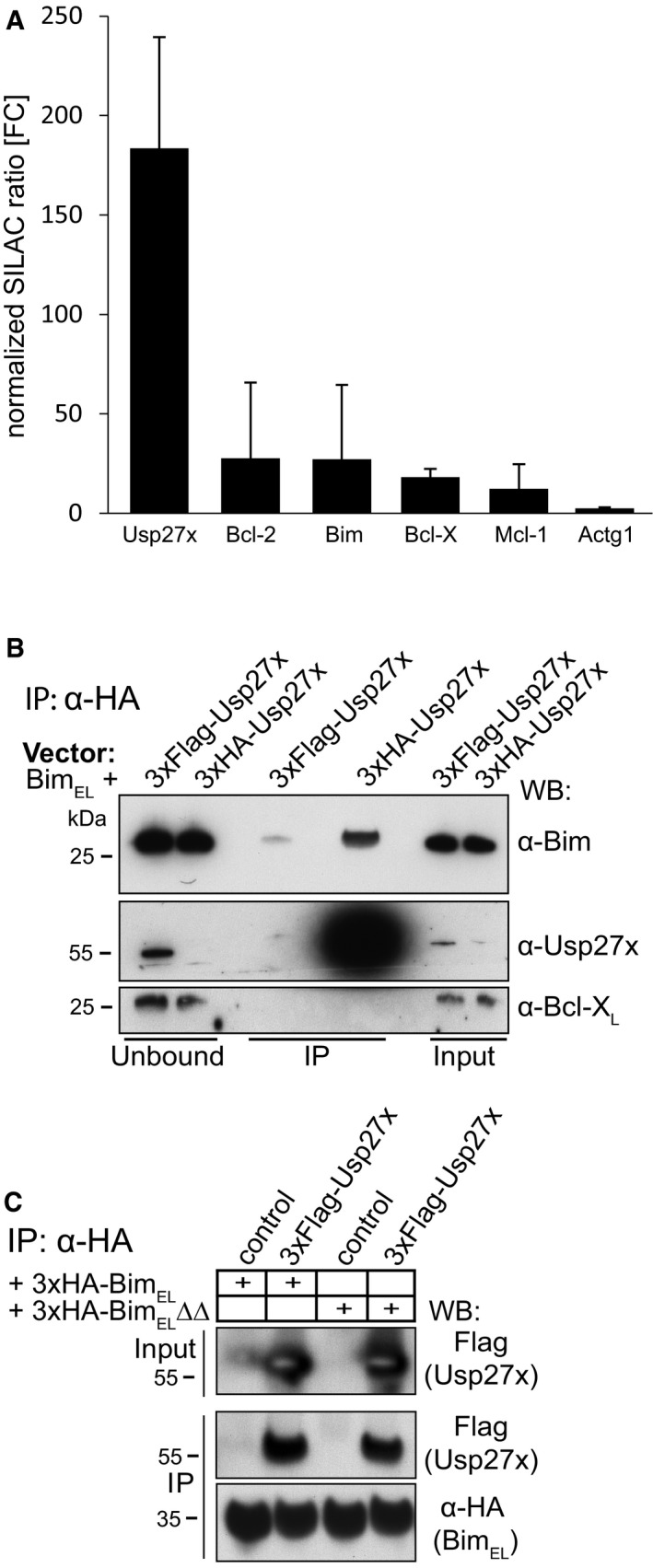
- Profile of enriched proteins in anti‐3xHA‐BIMEL immunoprecipitations of purified mitochondria from mouse embryonic fibroblasts (MEF Bax/Bak‐double knockout cells). Cells expressing 3xHA‐BimEL were labelled with light amino acids, whereas control cells (expressing only untagged BimEL) were labelled with heavy amino acids. Gamma actin (Actg1) served as a specificity control. Protein ratios are calculated from the respective peptide ratios. A minimum ratio count of two is needed to calculate a protein ratio. Error bars: protein quantification variability. Error bars indicate the coefficient of variability over all redundant quantifiable peptide signals. It is calculated as standard deviation of the fold enrichment of all peptide ratios (see also Table EV1). Identification of Usp27x as an interacting protein of BimEL was identified alongside the SILAC experiment published earlier. Enrichment of Bim and anti‐apoptotic proteins already appear in the earlier report 25.
- 293FT cells were transfected with expression constructs for untagged BimEL together with 3xFlag‐Usp27x or 3xHA‐Usp27x (all pMIG‐vector backbone). After cell lysis (1% Triton X‐100) and subsequent anti‐HA immunoprecipitation, the unbound fraction, input (50 μg each) and the IP‐eluate (IP, ˜60%) were run on SDS–PAGE. Flag‐ or HA‐tagged Usp27x were detected using an antibody directed against Usp27x (Abgent). See also Fig 1A. Similar results were seen in n = 3 experiments.
- Usp27x binds a mutant of Bim incapable of binding to anti‐apoptotic Bcl‐2 proteins. 293FT cells transfected with constructs encoding 3xFLAG‐Usp27x and 3xHA‐tagged BimEL (Fig 1B) or 3xHA‐tagged BimEL∆∆ (a mutant with two mutations in the BH3 domain, incapable of binding anti‐apoptotic Bcl‐2 proteins 50) were immunoprecipitated from whole‐cell extracts using anti‐HA resin. Bim and Usp27x were detected with anti‐HA and anti‐FLAG antibodies as indicated. Control, pEGFP‐N1 vector. The caspase inhibitor Q‐VD‐OPh (QVD) was added to the cultures to inhibit Bim‐induced apoptosis (see also Fig 1). Similar results were seen in n = 3 experiments.
