Figure EV3. Usp27x cleaves di‐ubiquitin molecules.
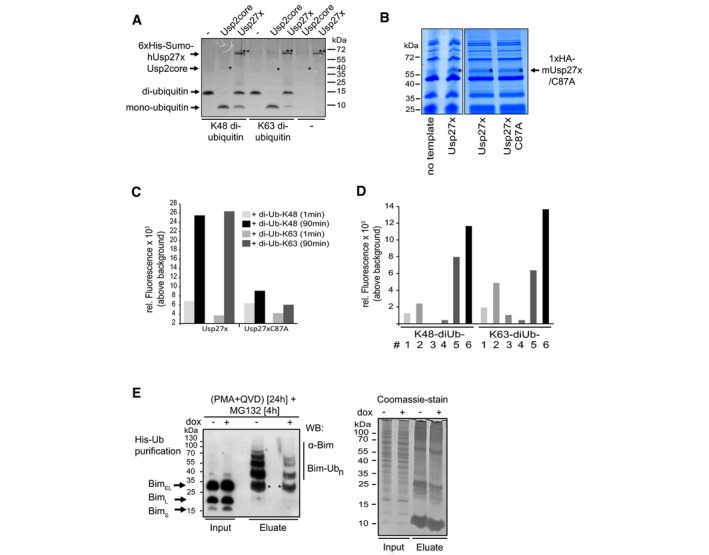
- Human 6xHis‐Sumo‐Usp27x was purified from E. coli. Differently linked di‐ubiquitin molecules (K48‐, K63‐linked di‐ubiquitin; 1 μg each) were incubated for 1 h at 37°C with 0.1 μM Usp2core (* positive control, ˜41 kDa) or 2.9 μM of the affinity‐purified 6xHis‐Sumo‐hUsp27x (**, ˜63 kDa). Usp27x was able to cleave K48‐ and K63‐linked di‐ubiquitin molecules into mono‐ubiquitin as visualized by Coomassie Blue staining. Similar results were observed in n = 2 separate experiments.
- Coomassie‐stained gels of soluble fractions (4 μl) from a PureExpress preparation without a template DNA (showing only proteins of the in vitro PureExpress translational E. coli system) or of in vitro‐translated 1xHA‐Usp27x (left gel) or the products of translation of 1xHA‐Usp27x and its catalytically inactive mutant 1xHA‐Usp27xC87A (right gel; Usp27x bands are marked with an asterisk). Reactions were used for (C). n = 2.
- Usp27x or Usp27xC87A were in vitro‐translated with the PureExpress system (see B). Four microlitres of soluble fractions of the reactions were directly used for assays of proteolytic activity. As substrates, K48‐linked IQF‐di‐UbK48‐6 or K63‐linked IQF‐di‐UbK63‐6 (200 nM each) (the 6 in K48‐6 and K63‐6 refers to the substrate variant where the quencher and TAMRA dye is linked at specific positions; these substrates were selected based on the preliminary experiment with the label at various positions (see Fig EV3D)) were added, and reactions were incubated for 1–90 min at 23°C. As readout for cleavage of K48/K63‐linked di‐ubiquitin, the relative fluorescence generated by dequenching was measured. Bars indicate mean values of duplicate measurements in one experiment.
- Four microlitres of soluble fractions (from the reactions shown in A) was directly used for the activity assay. Six different K48‐linked IQF‐diUb substrates and six K63‐linked IQF‐diUb substrates (substrates #1‐6; 200 nM each) were incubated with 1xHA‐Usp27x for 60 min at 23°C. As readout for cleavage of K48/K63‐linked di‐ubiquitin, the relative fluorescence was measured using a Tecan M200 fluorescence reader. IQF‐di‐UbK48‐6 and IQF‐di‐UbK63‐6 substrates showed the best performance and were further used for activity assays (see Fig EV3C). Bars indicate mean values of an experiment measured in duplicate.
- Usp27x expression reduces ubiquitination of Bim. 293FT‐TetR‐3xFlagUsp27x cells were transfected with a vector coding for 6His‐ubiquitin. After 24 h, cells were treated with PMA and QVD, and 3xFlag‐Usp27x was induced by dox at the same time. After additional 20 h, cells were treated for 4 h with MG132 (40 μM) to block proteasomal degradation of ubiquitinated proteins. Cells were lysed under denaturing conditions, and His‐ubiquitin‐labelled proteins were purified by Ni2+‐NTA affinity chromatography. Ubiquitinated Bim (Bim‐Ubn) was detected by Western blot using anti‐Bim antibodies (left). Asterisk (*) indicates non‐His‐ubiquitinated Bim that binds unspecifically to the Ni2+‐agarose beads (see Fig 2E). Equal loading was verified by a Coomassie‐stained SDS–PAGE from same protein extracts (right). Similar results were obtained in n = 3 separate experiments.
