Figure 1. Structural parameters of Toll-like receptor (TLR4) variants.
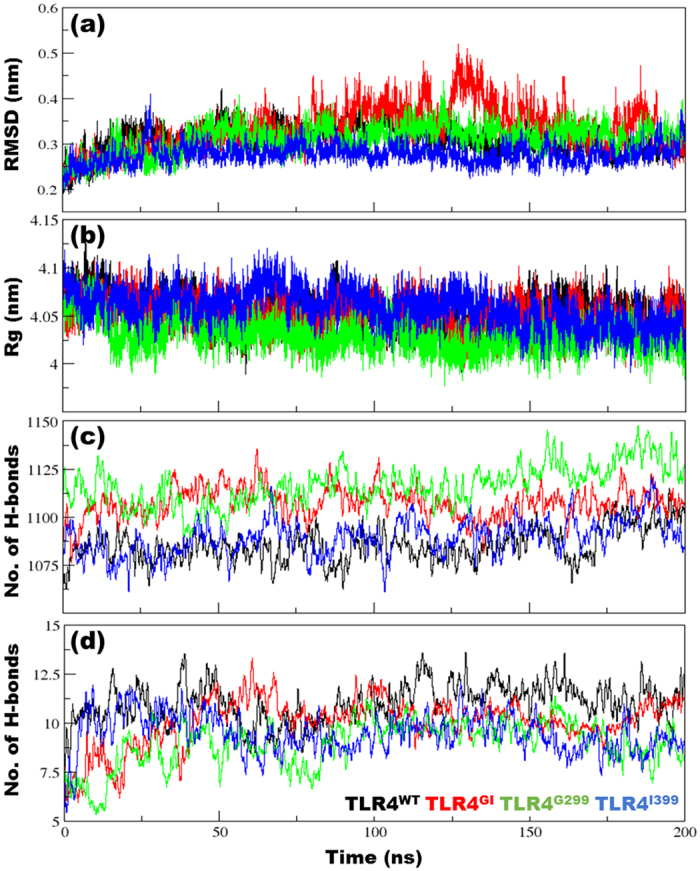
(a) Root mean-square deviation (RMSD) of backbone atoms after a least-squares fit to the initial structure over the whole trajectory. (b) Radius of gyration (Rg) of the backbone atoms in all complexes over the entire duration of simulation. (c) Number of intra-protein hydrogen bonds (H-bonds) and (d) protein-lipopolysaccharide (LPS) H-bonds for the entire duration of simulation. The criteria to measure H-bonds are based on cutoffs for the hydrogen-donor-acceptor angle (30°) and the donor-acceptor distance (0.35 nm). The OH and NH groups were regarded as donors, whereas O and N were acceptors.
