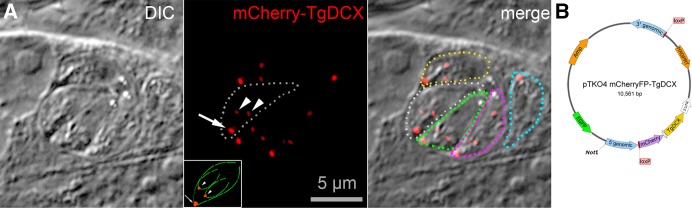
FIGURE 3:
Creation of a knock-in parasite line in which TgDCX was replaced with mCherryFP-TgDCX by homologous recombination. (A) Fluorescence and DIC images of mCherryFP-TgDCX knock-in parasites. The field of view includes one vacuole containing four parasites, each with two forming daughters, and another vacuole with one parasite with no daughter. The mCherryFP fluorescence is located in a single spot at the apical end of both adult (arrow) and developing daughter (arrowheads) parasites. For reference, a cartoon of a parasite with two daughters is included in the inset (see also Figure 1). One parasite of the four-parasite vacuole is enclosed by the dotted white line in the fluorescence and overlay panels. For clarity, the other four parasites are outlined in the overlay channel only (yellow, green, magenta, and cyan dotted lines). (B) Map of a TgDCX knock-in plasmid. Sandwiched between two LoxP sites are a selection marker (HXGPRT expression cassette), the coding sequence for mCherryFP-tagged TgDCX, and the 3′-UTR from the T. gondii GRA2 gene. Flanking the LoxP sites are 3′ and 5′ sequences, ∼900 base pairs each, copied from T. gondii genomic DNA that direct integration of the LoxP sandwich into the genome by homologous integration at the TgDCX locus, replacing the entire TgDCX coding region, exons, and introns. The plasmid is linearized by NotI cutting before electroporation into a Δhxgprt line of T. gondii (e.g., RHΔhx or RHΔku80Δhx). The EGFP expression cassette allows FACS or manual selection against nonhomologous recombinants. Integration at the TgDCX locus by homologous recombination with double crossover removes the region of the plasmid not framed by the 5′ and 3′ homology regions, including the EGFP expression cassette. After homologous recombination, expressing Cre recombinase in knock-in parasites deletes TgDCX and HXGPRT coding sequences from the genome (Figure 5).