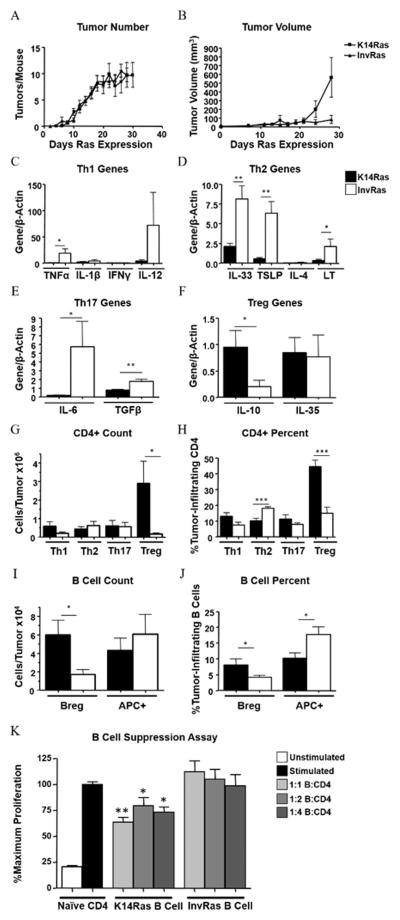
Figure 1. H-RasG12V expression in different epidermal layers determines distinct tumor phenotypes and immune responses.
Tumor counts (A) and tumor volume (B) in K14Ras and InvRas mice at indicated days after Ras induction. (A: K14Ras n = 45, InvRas n = 30. B: K14Ras n = 8, InvRas n = 10). (C–F) Relative expression of specific cytokines by qPCR in whole tumor tissue associated with Th1 (C), Th2 (D), Th17 (E), and Treg (F) cells. (C: K14Ras n = 8,16,5,15, InvRas n = 6,12,5,6. D: K14Ras n = 16,16,5,8, InvRas n = 12,12,5,6. E: K14Ras n = 8,16, InvRas n = 6,12. F: K14Ras n = 14,16, InvRas n = 10,10). Samples below detection level were not used in analysis. (G–J) Analysis of tumor-infiltrating CD4 T cell counts (G), and CD4 percentages (H), tumor-infiltrating B cell counts (I), and B cell percentages (J) in K14Ras and InvRas tumors. Counts were determined using a Cellometer Auto T4 Cell Viability Counter (Nexcelom Bioscience), and quantified using FlowJo software. Counts represent n = 12 for CD4 T cells, n = 11 for B cells. Percentages represent n ≥ 15 per group. (K) Analysis of CD4 T cell proliferation following 7 day coculture with tumor-conditioned B cells of K14Ras or InvRas mice. CFSE was analyzed by flow cytometry. Stimulated control baseline set to 100%, and significance is calculated against stimulated control group. n = 6 per group (2×3).