Figure 1. Sequence comparison between miR-26b and 3′UTR of CTGF and CCND1.
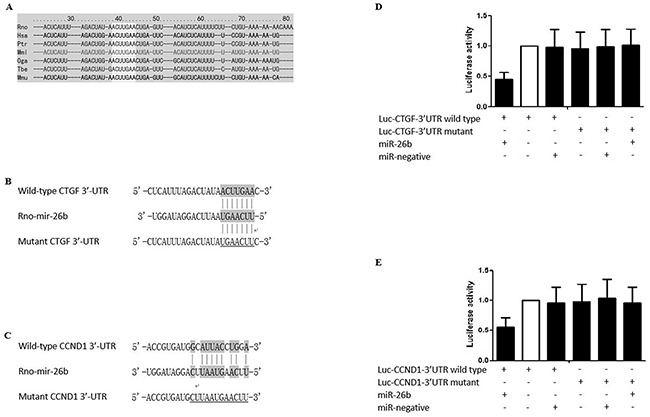
A. The “seed sequence” in the 3′ UTR of the target gene of rno-miR-26b is highly conserved among the species including, but not limited to, Rno, Has, Ptr, Mml, Oga, Tbe, and Mmu; B. Schematic comparison between rno-miR-26b and the wild-type (upper sequence)/mutated (lower sequence) 3′UTR of CTGF with “seed sequence” highlighted; C. Schematic comparison between rno-miR-26b and the wild-type (upper sequence)/mutated (lower sequence) 3′UTR of CCND1 with “seed sequence” highlighted; D. The relative luciferase activity in the rPASMCs transfected with both wild-type 3′UTR of CTGF and rno-miR-26b is significantly lower than the controls (p<0.01); E. The relative luciferase activity in the rPASMCs transfected with both wild-type 3′UTR of CCND1 and rno-miR-26b is significantly lower than the controls (p<0.01). All experiments were repeated three times (N value=3).
