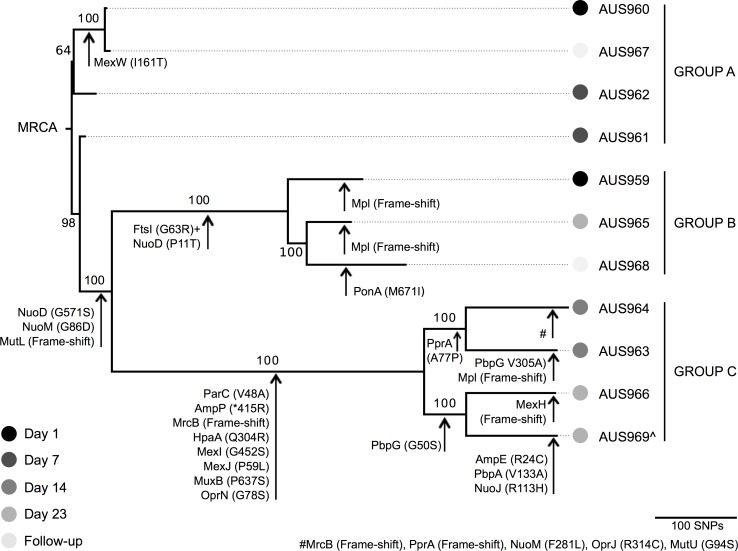
Fig 2. Phylogeny of M3L7 isolates from an individual with cystic fibrosis.
Isolates were cultured from sputum samples during intravenous treatment of an acute pulmonary exacerbation (Days 1, 7, 14 and 23) and at outpatient follow-up 8 weeks later. The phylogenetic tree was constructed based on a core SNP alignment of 1753 nucleotides generated from read mapping against PAO1. The genome of a M3L1 strain (isolate ID, AUS970), sequenced as part of an ongoing study, was used as an outgroup to root the tree. Isolates in Groups B and C are hypermutators whilst those in Group A are non-hypermutators. The scale bar represents 100 nucleotide substitutions. Amino acid changes (compared to PAO1) in genes associated with antibiotic resistance and hypermutation are indicated using an arrow. *Premature stop codon; +FtsI (G63C) identified in all other M3L7 isolates (Groups A and C); ^The M3L43 genotype (isolate ID, AUS969) is a derivative of the M3L7 sub-type; MRCA: most recent common ancestor. S2 Fig shows the phylogeny with predicted recombinant sites removed.