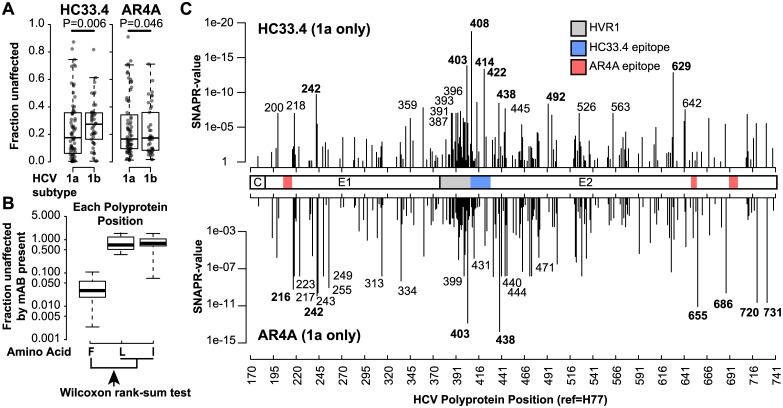
Fig 3. Subject-adjusted Neutralizing Antibody Prediction of Resistance-polymorphisms (SNAPR): Prediction of polymorphisms mediating neutralization resistance.
(A) Differences in HC33.4 and AR4A neutralization of HCVpp by subtype. Each data point indicates the mean Fu of an individual HCVpp, measured in duplicate. Boxes are interquartile range, and horizontal lines are medians. P-values calculated by Wilcoxon rank sum test. (B) Illustration of the SNAPR method: At each E1E2 alignment position, isolates were divided into groups according to amino acid present. Fraction unaffected (Fu, infection in the presence of bNAb/infection in the presence of nonspecific IgG) values of HCVpp from the most sensitive group (as determine by the medians) was compared to the Fu of HCVpp with any other amino acid at that position by Wilcoxon rank-sum test to generate SNAPR-values. (C) SNAPR-values across E1E2 determined using only subtype 1a HCVpp neutralized by HC33.4 or AR4A. The 8 positions with lowest SNAPR-values for HC33.4 and AR4A are in bold print. Previously defined HC33.4 and AR4A binding epitopes are indicated (blue and pink), as is hypervariable region 1 (HVR1) (gray).