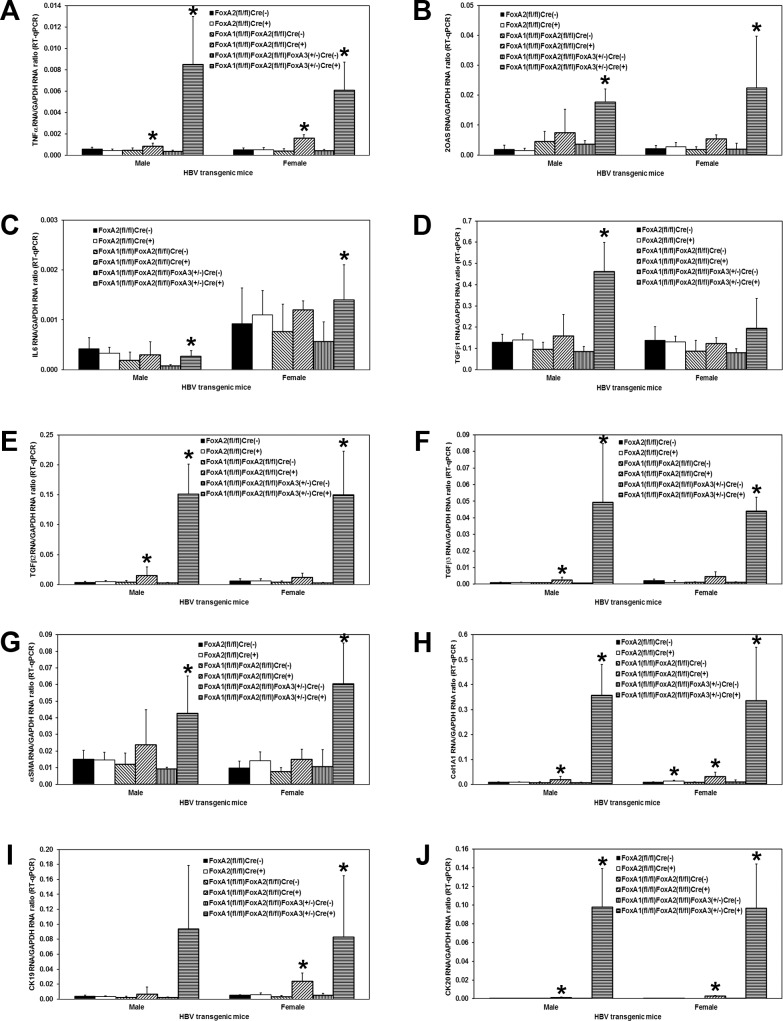
Fig 5. Effects of FoxA deletion on liver TNFα, 2OAS, IL6, TGFβ1, TGFβ2, TGFβ3, αSMA, Col1A1, CK19 and CK20 transcript levels in adult HBV transgenic mice.
Quantitative analysis of the (A) TNFα, (B) 2OAS, (C) IL6, (D) TGFβ1, (E) TGFβ2, (F) TGFβ3, (G) αSMA, (H) Col1A1, (I) CK19 and (J) CK20 transcripts by RT-qPCR in the HBV transgenic mice. The GAPDH transcript was used as an internal control for the quantitation of the TNFα, 2OAS, IL6, TGFβ1, TGFβ2, TGFβ3, αSMA, Col1A1, CK19 and CK20 RNAs. The mean relative TNFα, 2OAS, IL6, TGFβ1, TGFβ2, TGFβ3, αSMA, Col1A1, CK19 and CK20 transcript levels plus standard deviations derived from male and female FoxA-expressing (HBVFoxA2fl/flAlbCre(-), HBVFoxA1fl/flFoxA2fl/flAlbCre(-) and HBVFoxA1fl/flFoxA2fl/flFoxA3+/-AlbCre(-)) and FoxA-deleted (HBVFoxA2fl/flAlbCre(+), HBVFoxA1fl/flFoxA2fl/flAlbCre(+) and HBVFoxA1fl/flFoxA2fl/flFoxA3+/-AlbCre(+)) HBV transgenic mice is shown. The levels of the transcripts which are statistically significantly different between Cre(-) and Cre(+) HBV transgenic mice by a Student’s t-test (p<0.05) are indicated with an asterisk (*). Average number of mice per group was 6.5±1.8 (Range: 4–9).