Figure 4. ΔNp63 enhances CD44v6 expression.
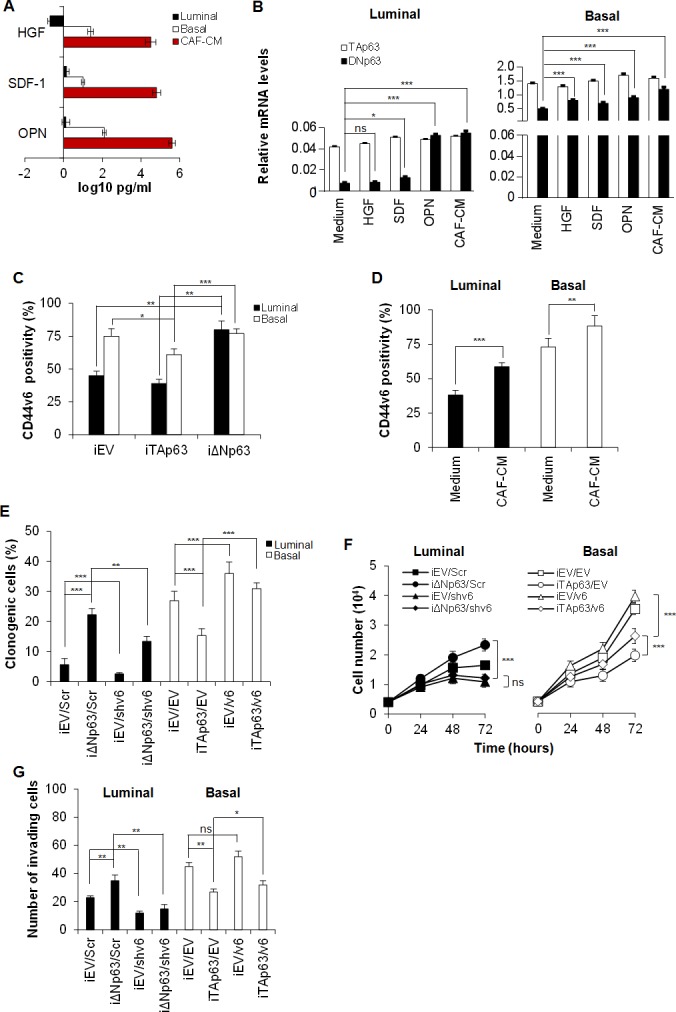
A. HGF, SDF-1 and OPN levels in supernatants of luminal and basal BCSCs, and cancer associated fibroblasts (CAFs) freshly isolated from a basal breast tumor sample. Data are mean ± SD of 3 independent experiments performed with 3 luminal and 2 basal BCSC lines derived from different patients. B. TAp63 and ΔNp63 mRNA expression levels in luminal and basal BCSCs, exposed to HGF, SDF-1 and OPN or CAF-conditioned medium (CAF-CM) for 24 hours. GAPDH amplification was used as endogenous control. Results show mean ± SD of 3 independent experiments performed with 3 luminal and 2 basal BCSC primary lines. C. Percentage of CD44v6 positive cells in luminal and basal iEV, iTAp63 or iΔNp63 BCSCs performed by flow cytometry. Data are shown as mean ± SD of 3 independent experiments using 3 luminal and 2 basal BCSC primary lines. D. CD44v6 positivity percentage in luminal and basal BCSCs treated with medium (Medium) or CAF-CM for 24 hours. Results are mean ± SD of 3 independent experiments accomplished using 3 luminal and 2 basal BCSCs derived from different patients. E. Percentage of clonogenic luminal BCSCs transduced with either or both iΔNp63 and shCD44v6 (shv6) vector and their control, iEV and Scr respectively. Basal BCSCs were transduced with either or both iTAp63 and CD44v6 (v6) expressing vectors and their iEV/EV controls. BCSCs were firstly transduced with the inducible isoforms of p63 and after one week with shv6 and v6. After 7 days, cells were exposed to doxycycline for 5 days before the experiment and plated in doxycycline containing medium for the entire assay. Results are mean ± SD of 3 independent experiments performed with 3 luminal and 2 basal BCSC primary lines. F. Cell number analysis of luminal and basal BCSCs as in E, up to 72 hours. G. Invasion assay at 72 hours of luminal and basal BCSCs treated as in E. Results are mean ± SD of 4 independent experiments. * indicates P < 0.05, ** indicate P < 0.01 and *** indicate P < 0.001. ns indicates non statistically significant.
