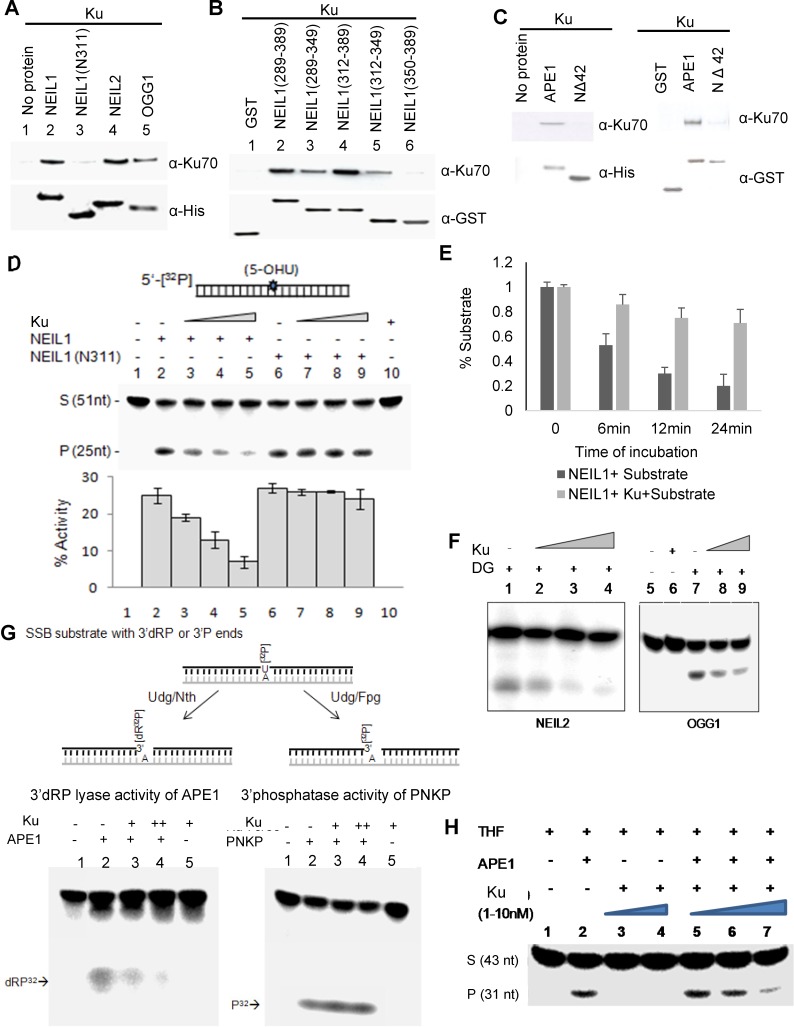
Figure 2. Ku inhibits DGs direct interaction.
A. Direct interaction of Ku with NEIL1 (but not the N311 mutant), NEIL2, and OGG1 was confirmed by His-affinity pull down analysis using purified proteins in the absence of DNA. B. GST pull down analysis showing that NEIL1 aa289-349, but not aa350-389, was needed for Ku binding. C. The binary interaction of Ku with APE1 (but not the NΔ42 mutant) is shown by His-affinity pull down analysis. D. Dose-dependent inhibition of WT NEIL1 (1nM, lane 2) by Ku (0.1, 0.5 and 1nM; lanes 3-5), but not by the N311 mutant (1nM, lane 6-9) is shown with a 5-OHU-containing 51-mer duplex substrate (25nM). E. Time kinetics of Ku inhibition of NEIL1 activity. F. NEIL2 and OGG1 are also inhibited by Ku at similar experimental conditions. G. A 26-nt oligo containing U at the 5′ terminus was labelled with γ32P-ATP using T4-PNK and annealed with a 25-nt proximal sequence and a 51-nt complementary oligo. The duplex was digested with Udg/Nth or Udg/Fpg to generate a strand break with a 3′dRP or 3′P end, respectively (top). Ku inhibits 3′dRPase activity of APE1 (bottom left panel), but not the 3′phosphatase activity of PNKP (bottom right panel). H. Ku inhibition of AP endonuclease activity of APE1.