Abstract
Long non-coding RNAs (lncRNAs) are associated with the occurrence, development and prognoses of non-small cell lung cancer (NSCLC). In the present study, we investigated the functional mechanisms of the lncRNA XIST in two human NSCLC cell lines, A549 and NCI-H1299. In all the 5 NSCLC cell lines (NL9980, NCI-H1299, NCI-H460, SPC-A-1 and A549) tested, the expression levels of XIST were significantly elevated, as compared with those in normal human bronchial epithelial cell line BEAS-2B. In A549 and NCI-H1299 cells, knockdown of XIST by siRNA significantly inhibited the cell proliferation, migration and invasion, and promoted cell apoptosis. Furthermore, XIST knockdown elevated the expression of E-cadherin, and suppressed the expression of Bcl-2. Moreover, knockdown of XIST significantly suppressed the tumor growth in NSCLC A549 xenograft mouse model. Bioinformatic analysis and luciferase reporter assays revealed that XIST was negatively regulated by miR-449a. We further identified reciprocal repression between XIST and miR-449a, which eventually influenced the expression of Bcl-2: XIST functioned as a miRNA sponge of miR-449a, which was a negative regulator of Bcl-2. These data show that expression of the lncRNA XIST is associated with an increased growth rate and metastatic potential in NSCLC A549 and NCI-H1299 cells partially through miR-449a, and suggest that XIST may be a potential prognostic factor and therapeutic target for patients with NSCLC.
Keywords: non-small cell lung cancer, A549 cells, NCI-H1299 cells, A549 xenografts, lncRNA, XIST, miR-449a, tumor progression
Introduction
Lung cancer is a predominant cause of cancer-related death worldwide due to its high morbidity and lack of effective therapy strategies1. Almost 80% of lung cancers are non-small cell lung cancers (NSCLCs)2. Despite recent advancements in clinical and experimental oncology, the prognosis of patients with NSCLC remains poor, with a 5-year overall survival rate of approximately 11%3. Treatment failure and death of patients with NSCLC are correlated with a high potential for invasion and metastasis. Therefore, understanding the mechanisms underlying NSCLC development and progression is essential to improve the diagnosis, prevention, and treatment of NSCLC patients.
Accumulating data from whole genome and transcriptome studies have revealed that only a small percentage (1%–2%) of the genome encodes proteins4,5, and the majority of the mammalian genome encodes vast numbers of non-coding RNAs6. Long non-coding RNAs (lncRNAs) are a novel class of transcripts with no protein-coding capacity but with diverse functions in cancer cell proliferation, apoptosis and metastasis. Recent studies have indicated that the type and number of lncRNAs vary greatly among species, tissues and cells7,8,9, and these macromolecules are biologically functional and associated with the development, progression and treatment response of various diseases, including cancer10. Previous studies have shown that MALAT111, HOTAIR, H1912, the lncRNA GAS6-AS113 and SCAL114 are associated with the development and progression of lung cancer. However, the functional mechanism of lncRNAs remains largely unknown.
XIST (X-inactive specific transcript) is an lncRNA required for the X chromosome inactivation (XCI) of one of the two X chromosomes in females and XY males15. Considerable evidence has shown that XIST plays important roles in the differentiation, proliferation and genome maintenance of human cells. Moreover, XIST has been found to be dysregulated in a variety of human cancers, and this dysfunctional expression has been shown to exert a pathological role16. Recent studies have suggested that the expression of XIST is associated with glioma stem cells (GSCs) in human glioma17. However, the function of XIST in lung cancer, particularly in NSCLC, remains a mystery.
In the present study, we investigated the expression and function of XIST in NSCLC. First, we observed remarkably upregulated XIST expression levels in NSCLC cells. Knockdown of XIST suppressed NSCLC cell growth and metastasis, implying a possible role of lncRNA as an oncogene in lung cancer. Moreover, we found that miR-449a interacts with XIST by directly targeting the miRNA-binding site in the XIST sequence, and XIST may be a ceRNA of miR-449a.
Methods and materials
Cell culture and transfection
All cells were obtained from the Tianjin Lung Cancer Institute and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin in humidified atmosphere containing 5% CO2 at 37 °C. Transfection of either miRNAs or siRNAs was conducted using Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer's recommendations. The mature hsa-miR-449a mimic/inhibitor, siRNA targeting XIST, lentivirus, and corresponding negative controls were all purchased from GenePharma (Shanghai, China). The siRNA sequences are listed in Table S1.
Quantitative real-time PCR
Quantitative real-time PCR was performed using SYBR Premix Ex Taq™ (Takara, Japan). Total RNA isolated using TRIzol reagent (Invitrogen, CA, USA) was subsequently reverse-transcribed to cDNA according to the manufacturer's protocol. U6 and GAPDH were used as internal controls for non-coding RNA and mRNA, respectively. All primer sequences are listed in Table 1. The reactions were aliquoted in either a 96-well plate (ABI) or a 384-well plate and conducted using a pre-heated real-time PCR instrument (ABI 7500 or ABI 7900, respectively). The PCR program consisted of 10 s at 95 °C followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. Three independent experiments were performed to analyze relative gene expressions and each sample was tested with technical triplicates or quadruplicates. Ct values were used to calculate the RNA expression levels. The target gene expression based on the 2−ΔΔCt method was normalized using endogenous U6 or GAPDH as references, and the amount of target gene in the control sample was set at 1.0.
Target prediction
The target genes of XIST and their relationship with miR-449a were predicted using three bioinformatics algorithms: TargetScanS, miRanda (microrna.org) and starBase V2.0 (starbase.sysu.edu.cn).
Plasmid construction and luciferase reporter assay
The mutated miR-449a binding sites within the XIST luciferase reporter vector were generated by site-directed mutagenesis. The XIST gene was amplified from human genomic DNA, and the PCR product was cloned into the pMIR-GLO™ Luciferase vector (Invitrogen, USA) downstream of the firefly luciferase coding region. The construct was named pMIR-XIST-wt. Mutations of the miR-449a binding sites were introduced by site-directed mutagenesis, and the resulting plasmid was referred to as pMIR-XIST-Mut. All of the sequences of generated reporter plasmids were verified by sequence analysis. Luciferase assays were conducted using 1×104 H1299 cells plated in a 48-well plate. Co-transfection was performed using 200 ng of either pMIR-XIST-wt or pMIR-XIST-Mut vector and 80 ng of either miR-449a mimics or inhibitor. At 48 h after transfection, the cells were harvested and assayed for both firefly and Renilla luciferase using the Dual-Glo Luciferase Assay Kit (Promega, Madison, WI, USA). All transfection experiments were conducted in triplicate.
Cell migration and invasion assay
Wound healing assays were used to measure cell migration capability. In 6-well plates, XIST siRNAs (siXIST) and negative controls (NC) were transiently transfected into A549 and H1299 cells. After 24 h, when cells grew to 90% confluency, cell monolayers were scratched with a sterile micropipette tip. In the next step, wounded monolayers were washed with phosphate-buffered solution (PBS) to remove cell debris. At two time points, the distance between the two edges of the wound was calculated at three different positions. In addition, cells were measured every 24 h. The Transwell assay was used to measure the capabilities of cell migration (without Matrigel in the upper chambers) and invasion (with Matrigel in the upper chambers). A549 and H1299 cells were transfected with either siXIST or NC. Approximately 16 h later, transfected cells were trypsinized and resuspended, and 5.0×103 cells in 200 μL of DMEM were placed into the upper chambers (8-mm pore size; Millipore, USA). The lower chambers were filled with 600 μL complete medium with 10% FBS. After a 48-h incubation at 37°C, non-invading cells were removed from the top of the chamber with a cotton swab. The invaded cells on the lower surface of the inserts were fixed and stained with 0.1% crystal violet, and cells were counted in five random fields for each insert at 200× magnification. Transwell assays were conducted in duplicate in two separate experiments. The QCM™ 24-Well Cell Invasion Assay (Fluorometric) (Millipore, USA) was conducted according to the manufacturer's recommendation.
Cell counting Kit-8 (CCK-8) assay
Transfected and untreated cultured cells (2.5×103) were seeded into 96-well plates (Corning Inc, USA). Cell proliferation was assessed at different time points (0, 24, 48, 72, and 96 h), using the CCK-8 assay (Dojindo, Tokyo, Japan) according to the manufacturer's protocol. The absorbance value at a wavelength of 450 nm, which is indicative of a positive relationship with cellular proliferation, was measured using a microplate reader (Molecular Devices, USA).
Colony formation assay
For the colony formation assay, cells were seeded at a density of 500 cells per well in 6-well plates, cultured for 7 or 12 d, fixed with a 4% paraformaldehyde solution, stained with 0.5% crystal violet and counted under an inverted microscope.
Anchorage-independent soft-agar growth assay
For the anchorage-independent soft-agar growth assay, transfected A549 and H1299 cells (4×104) were suspended in 1 mL of complete medium containing 0.66% agar (Sigma, Japan) and then applied over a layer of 1% agar/complete medium in 6-well plates. After 7 or 12 d, the number of colonies was counted under an inverted microscope.
Cell apoptosis assay
Cell apoptosis was measured by flow cytometry analysis following the instructions of the Annexin V-fluorescein isothiocyanate (FITC)/PI Apoptosis Detection Kit (R&D Systems, Abingdon, UK). Cultured cells were washed twice in PBS, suspended in 500 mL binding buffer, and labeled with 5 mL annexin V-FITC/PI for 15 min in the dark at room temperature. Afterwards, flow cytometry was performed on a FACSAria flow cytometer (BD Biosciences, USA). The data were analyzed using BD FACSDiva software.
In vivo tumorigenesis
Male BALB/c nude mice (4 weeks old) were purchased from the Charles River Company in China. Lentivirus was purchased from GenePharma (sequence shown in Supplementary Table 2). Screening of stably transfected A549 cells were seeded into 6-well plates (2×106 cells/well) in antibiotic-free media, and the cells were then subcutaneously injected into the dorsal flank of nude mice (1×106 cells/mouse, n=5). Tumor diameters were measured every 5 d, and the tumor volumes were calculated using the equation: volume=D×d2×π/6, where D and d represent the longest and shortest diameters, respectively. All animal handling and research protocols were approved by the Animal Care and Use Ethnic Committee of Tianjin Medical University.
Western blot
Proteins were separated on 10% or 12% SDS-PAGE gels and transferred onto polyvinylidene fluoride membranes (Millipore, USA); the membranes were then blocked with 5% non-fat dry milk in Tris-buffered saline with Tween-20 (TBS-T), incubated with either rabbit or mouse primary antibody (1: 1000; CST, USA) at 4 °C overnight, and incubated with anti-rabbit or anti-mouse IgG secondary antibody (1: 2000; Beyotime, CA, USA). The bands were detected by enhanced chemiluminescence (Immobilon ECL, Millipore, USA), and β-actin (1: 5000; CST, USA) was used as a loading control.
Statistical analysis
SPSS 13.0 software (IBM SPSS Inc, USA) was applied to perform the data processing. Each experiment was repeated at least three times. Statistical significance was assessed by comparing the mean±SD using Student's t-test for independent groups. The results were considered statistically significant when P<0.05 and P<0.01.
Results
LncRNA XIST is upregulated in lung cancer cells
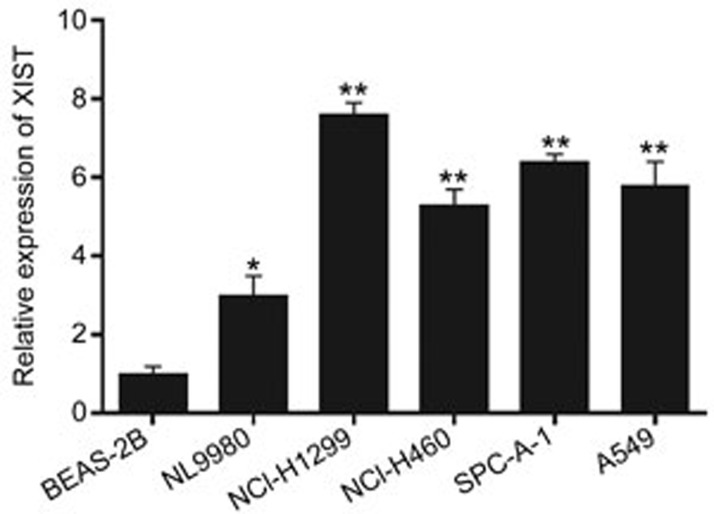
The expression of XIST in the NSCLC cell lines (NL9980, NCI-H1299, NCI-H460, SPC-A-1, and A549) was detected by qRT-PCR. Compared with human normal bronchial epithelial BEAS-2B cells, the expression of XIST was increased in all five NSCLC cell lines (Figure 1). The results suggested that the lncRNA XIST might play a critical role in the progression of NSCLC.
Figure 1.
Expression of XIST in NSCLC cells. Detection of XIST expression in lung cancer cells and a human normal bronchial epithelial cells using real-time PCR. Each treatment was performed with at least three biological replicates. Mean±SD. *P<0.05, **P<0.01.
Knockdown of XIST inhibits proliferation and induces apoptosis of NSCLC cells
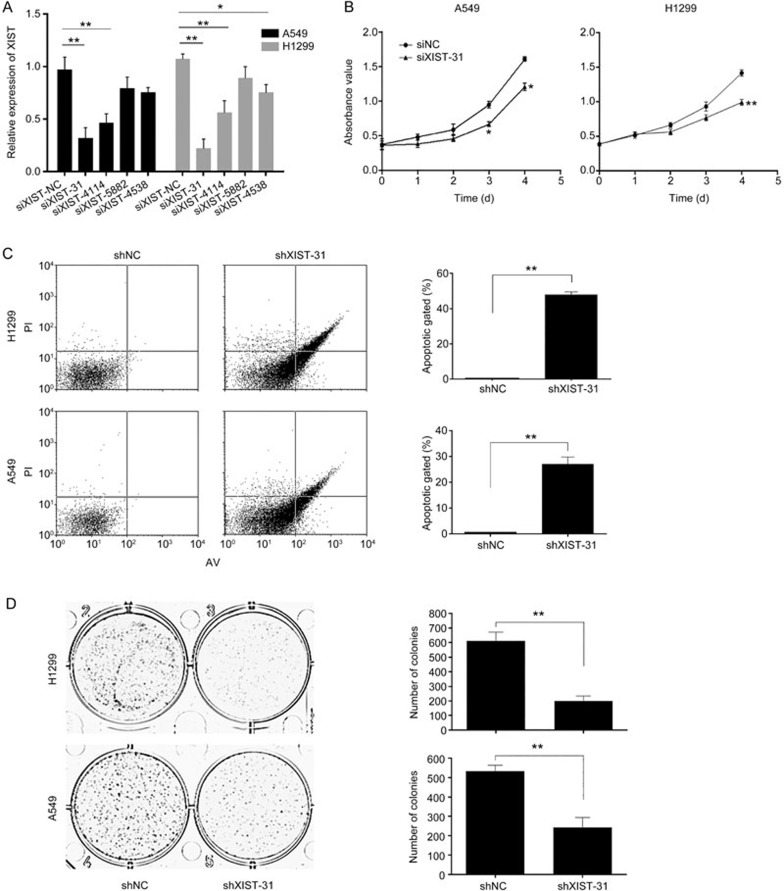
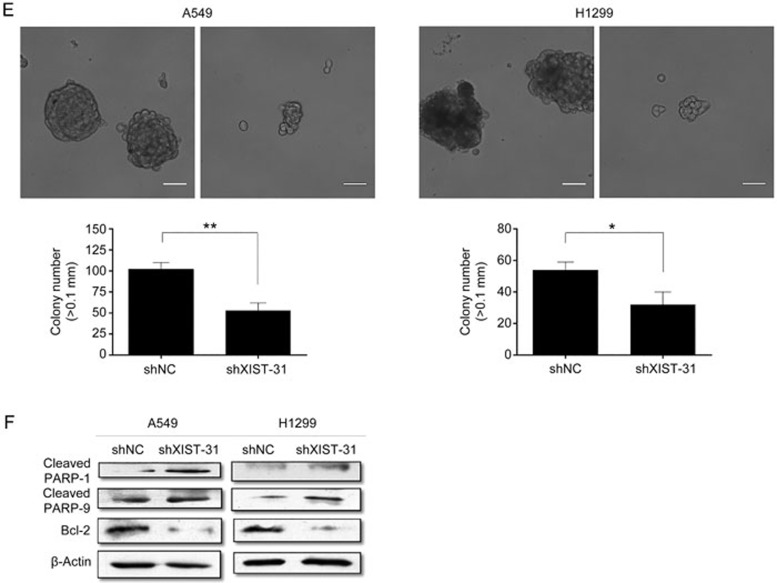
A previous study showed that XIST could associate with the occurrence and development of various cancers, including lung cancer. Hence, we explored how XIST affects lung cancer cells by next investigating its biological effects. We downregulated XIST in A549 and H1299 cells using short interfering RNAs (siRNAs), and four siRNAs as well as a negative control were designed according to different targets. At 48 h post-transfection, XIST was clearly reduced by siXIST-31 and siXIST-4114 in both A549 and H1299 cells. The knockdown effect of siXIST-31 was better than that of the other siRNAs, so we used siXIST-31 for all subsequent experiments (Figure 2A). To investigate the function of XIST on the proliferation of A549 and H1299 cells, the CCK-8 assay was performed. The OD450 value in the CCK-8 assay revealed that knockdown of XIST inhibited A549 and H1299 cell proliferation compared with that of the blank control group (Figure 2B). Annexin V staining showed that the percentage of early apoptotic cells following knockdown of XIST was drastically increased relative to that in control groups (Figure 2C). The results of the colony formation assay revealed that clonogenic survival of A549 and H1299 cells was decreased following knockdown of XIST (Figure 2D). Similarly, knockdown of XIST significantly suppressed the anchorage-independent colony formation of both A549 and H1299 cells (Figure 2E). Finally, based on the apoptotic results in Figure 2C, we detected the protein levels of classic apoptotic markers in A549 and H1299 cells. Among these markers, cleaved PARP-1, cleaved caspase-9, and Bcl-2 showed differential expression levels that were consistent with a change in the apoptosis rate (Figure 2F).
Figure 2A–2D.
Effect of XIST knockdown on the growth and apoptosis of lung cancer cells. (A) We designed siRNA for different gene loci of XIST gene and detected the knockdown effect by real-time PCR. (B) Growth curves of A549 and H1299 cells after transfection with siXIST or siNC were determined by CCK8 assays. (C) The effect of XIST knockdown on apoptosis in A549 and H1299 cells was determined by measuring the percentage of Annexin V-stained cells using flow cytometry. (D) Representative images (left) and quantification (right) of the colony formation assay of A549 and H1299 cells transfected with siXIST or siNC. Mean±SD. *P<0.05, **P<0.01.
Figure 2E, 2F.
(E) Soft agar colony formation assays were performed to determine the effect of XIST knockdown on the anchorage-independent growth of A549 and H1299 cells. (F) Effect of siXIST or siNC on the expression of PARP-1, caspase-9 and Bcl-2 in transfected A549 and H1299 cells. Each treatment was performed with at least three biological replicates. Mean±SD. *P<0.05, **P<0.01.
Knockdown of XIST inhibits migration and invasion of NSCLC cells
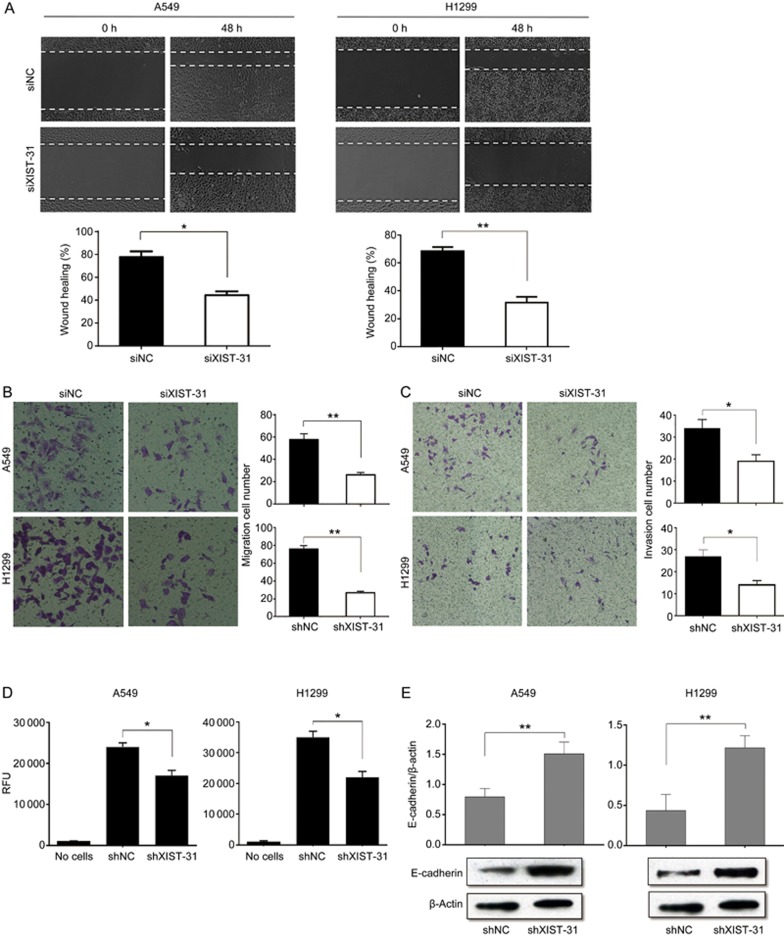
Cell migration and invasion are significant aspects of cancer progression that involve the migration of tumor cells into contiguous tissues and the dissolution of extracellular matrix proteins. Here, we evaluated cancer cell migration by using the wound scratch assay, transwell assay, and fluorescence-based transwell assay. After transfection with either XIST siRNA or negative control, significant differences were observed in cell migration. When compared with the negative control, the migratory ability of A549 and H1299 cells was decreased in the siXIST group (Figure 3A). Moreover, cell migration as determined by the transwell assay without Matrigel was decreased in siXIST-transfected A549 and H1299 cells (Figure 3B). The inverted fluorescence microscopy counting method and fluorescence detection format were used in the transwell assay with Matrigel to evaluate the cell invasion ability. Coincidentally, A549 and H1299 cells infected with lentivirus that expressed shXIST showed reduced cell invasion ability compared with cells infected with negative control shRNA sequences (Figure 3C, 3D). Western blot was performed to verify the protein levels of E-cadherin in A549 and H1299 cells as well as to further explore the effect of EMT. The results indicated that knockdown of XIST induced E-cadherin protein expression (Figure 3E). These results suggested that knockdown of XIST had a negative effect on cell migration and invasion in NSCLC.
Figure 3.
Effect of knockdown of XIST gene on cell metastasis. (A) Wound healing assay was used to demonstrate the effects of XIST knockdown on migration of A549 and H1299 in vitro. (B, C) Transwell assay was used to detect cell migration and invasion with knockdown of XIST. (D) Fluorescence detection format were used in transwell assay with matrigel to evaluate the cell invasion ability. (E) Effect of XIST knockdown on expression of E-cadherin detected by Western blot in A549 and H1299 cells. Each treatment was performed with at least three biological replicates. Mean±SD. *P<0.05, **P<0.01.
Knockdown of XIST inhibits tumor growth in vivo
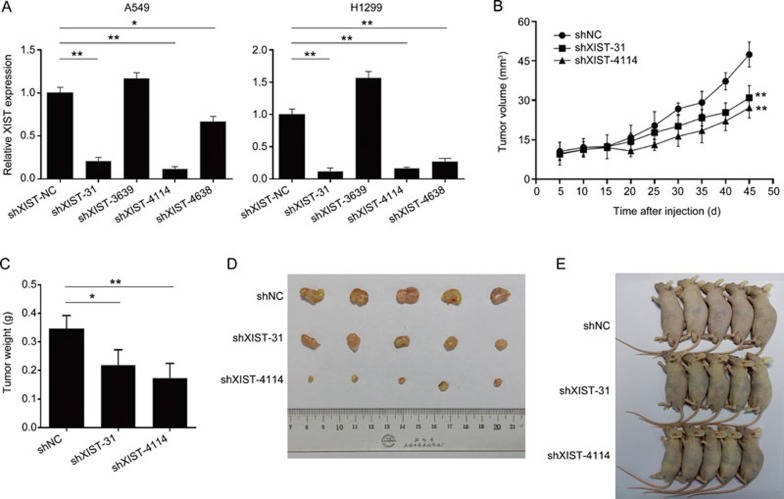
To explore whether the lncRNA XIST affects tumorigenesis, A549 cells were infected with the lentivirus containing shXIST and injected into a nude mouse to produce a xenograft model. Because of the improved knockdown efficiency of shXIST-31 and shXIST-4114 (Figure 4A), A549 cells were infected with lentivirus containing shXIST-NC, shXIST-31 and shXIST-4114. At 45 days after injection, a dramatic decrease in the tumor volume and weight were observed in the shXIST-31 and shXIST-4114 groups compared with the shXIST-NC group (Figure 4B–4E).
Figure 4.
Effect of XIST knockdown on the tumor growth in vivo. (A) We designed lenti-virus for different gene loci of XIST gene and detected the knockdown effect by real-time PCR. (B) Tumor volumes were calculated after injection every five days. (C) Tumor weight after XIST gene was knocked down. (D, E) Empty vector shNC, shXIST-31 and shXIST-4114 was transfected into A549 cells, which were injected in the male nude mice (n=5), respectively. Each treatment was performed with at least three biological replicates. Mean±SD. *P<0.05, **P<0.01.
XIST is negatively regulated by miR-449a
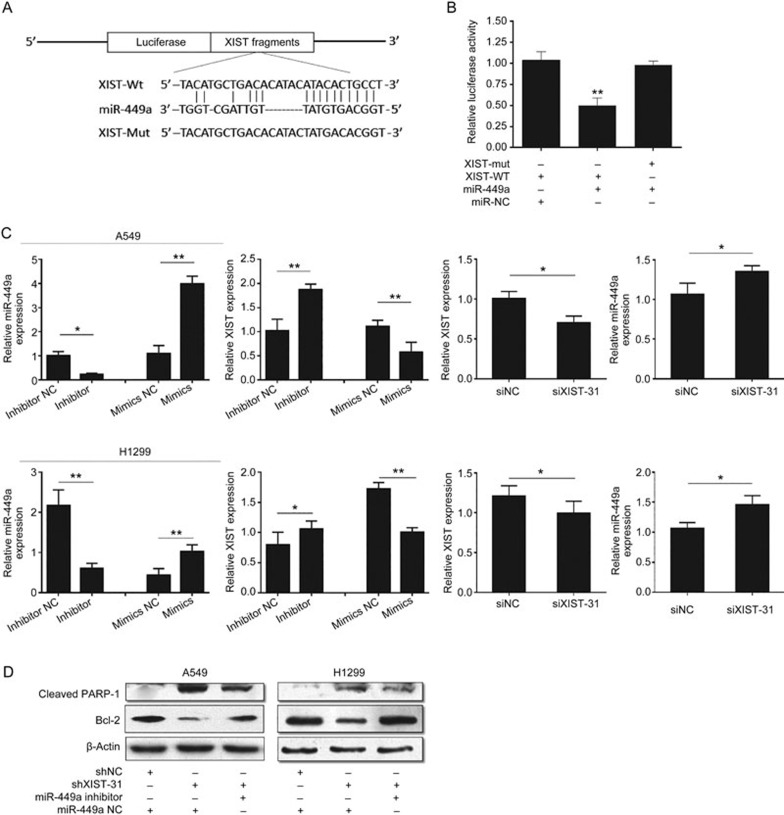
Our previous study showed that miR-449a may function as a tumor suppressor in lung cancer18. Bioinformatics analysis of miRNA recognition sequences on XIST revealed the presence of miR-449a tumor-suppressive binding sites (Figure 5A). To further confirm that the reduction in relative luciferase activity from the Luc-XIST-WT vector was due to a direct interaction between miR-449a and its putative binding site, we mutated the miR-449a binding site by site-directed mutagenesis, which resulted in Luc-XIST-Mut. As expected, the suppression of luciferase activity was completely abolished in this mutant construct compared with that in the wild-type XIST construct (Figure 5B). A549 and H1299 cells were transfected with siXIST, miR-449a inhibitor or miR-449a mimics. Then, the expression levels of miR-449a and XIST were detected by quantitative real-time PCR. The results showed reciprocal repression of XIST and miR-449a (Figure 5C). To further confirm the cross-talk between XIST and miR-449a, we co-transfected shXIST and a miR-449a inhibitor and measured the protein levels of cleaved PARP-1 and Bcl-2. The results showed that knockdown of XIST increased cleaved PARP-1 and suppressed Bcl-2 protein levels in both A549 and H1299 cells. Moreover, cells transfected with either miR-449a-NC or the miR-449a inhibitor could partly reverse the observed changes in the Bcl-2 and PARP-1 protein levels (Figure 5D).
Figure 5.
The interaction between XIST and miR-449a is regulated by each other. (A) Prediction consequential pairing of target region between XIST and miR-449a by bioinformatics method and construction of reporter gene plasmids and point mutation plasmids. (B) Luciferase reporter assay results demonstrated miR-449A can decrease the activity of wild-type gene plasmid XIST report. (C) Real-time PCR was used to detect the association between XIST and miR-449a. (D) The expression of PARP-1 and Bcl-2 in A549 and H1299 cells transfected with shNC, shXIST, miR-449a inhibitor or miR-449a NC by Western blot. Each treatment was performed with at least three biological replicates. Mean±SD. *P<0.05, **P<0.01.
Overexpression of XIST promotes cell migration and invasion by miR-449a inhibitor
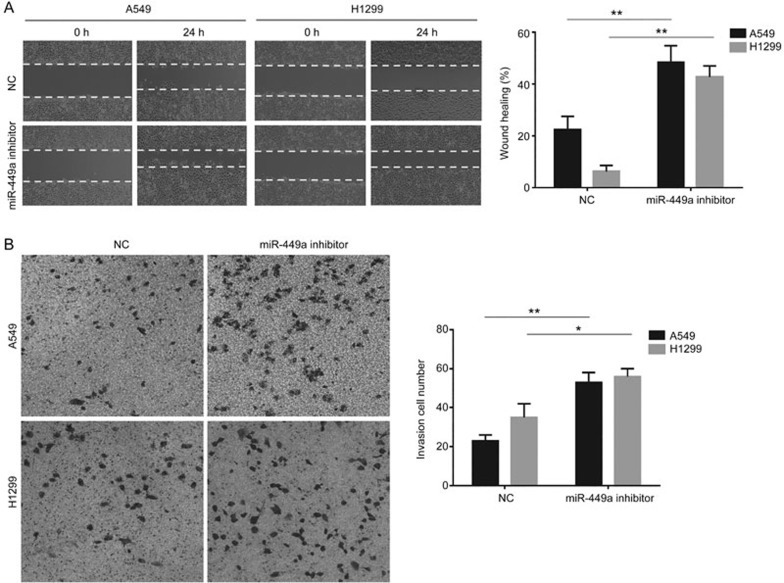
A previous study showed that reduced expression of miR-449a may promote XIST expression. Therefore, we induced overexpression of XIST treating cells with a miR-449a inhibitor and subjecting them to cell migration and invasion assays. The wound healing results revealed that XIST upregulation promoted cell migration in A549 and H1299 cells (Figure 6A). Additionally, the transwell assay showed an enhanced invasive ability in both A549 and H1299 cells when XIST was overexpressed (Figure 6B).
Figure 6.
Effects of upregulation of XIST gene expression on cell metastasis. (A) Wound healing assay was used to detect the effect of upregulation of XIST on migration of A549 and H1299 in vitro. (B) Effects of upregulated XIST on invasion of A549 and H1299 detected by transwell assay in vitro. Each treatment was performed with at least three biological replicates. Mean±SD. *P<0.05, **P<0.01.
Discussion
Lung cancer is the most common cause of cancer-related death worldwide. The prognosis for patients with NSCLC is still dismal, and the median survival of patients with untreated NSCLC is only four to five months. Recently, more studies have revealed that lncRNAs play important roles in the development and progression of multiple cancers, including NSCLC19,20. For example, knockdown of the lncRNA TATDN1 has been shown to inhibit tumor growth and metastasis in NSCLC cells by inhibiting β-catenin and Ezrin; thus, TATDN1 may be a potential prognostic factor and therapeutic target for NSCLC21. In addition, the lncRNA ANRIL has been found to be significantly upregulated in NSCLC tissues, and its expression promotes cell proliferation and inhibits cell apoptosis in vitro and in vivo22. Moreover, HOTAIR, MALAT1 and SBF2-AS1 have been shown to exert oncogenic functions in NSCLC23,24,25. However, even in light of these discoveries, other NSCLC-related lncRNAs and their modulatory effects have not yet been identified.
The full lncRNA XIST gene sequence is 32103 bp, so it is difficult to synthesize an effective overexpressing sequence of the XIST gene. Therefore, knockdown of XIST was performed in our study to explore the biological function of XIST. A previous study showed that XIST downregulation exerts tumor-suppressive functions by reducing cell proliferation, migration, and invasion as well as inducing apoptosis in GSCs17. While XIST has been reported to be a potent suppressor of hematological cancer26, our results revealed that knockdown of XIST promotes NSCLC cell apoptosis and inhibits cell proliferation, migration and invasion. The in vivo studies demonstrated that XIST downregulation suppresses tumor growth. All of these data indicate that XIST plays an important role in the development of NSCLC. However, XIST repression may hinder pluripotent stem cell induction at early stages in women27 and might be critical for the long-term survival of stem cells26. Hence, XIST is associated with the evolution of cancer stem cells to tumor cells.
It is widely known that mammalian genomes encode thousands of lncRNAs in addition to protein-coding RNAs28. These lncRNAs do not encode proteins but rather interact with other ncRNAs such as miRNAs as well as with each other. Many complex regulatory networks are formed by lncRNAs to regulate the expression of protein-coding genes at different levels. For example, HOTAIR may work as a regulator of miR-326 and then affect the targeted protein expression of Phox2a, which subsequently regulates cell proliferation and migration in lung cancer29. The lncRNA PCAT-1 promotes cancer cell proliferation through c-Myc30, and the lncRNA LUCAT1 is associated with poor prognosis in human NSCLC and affects cell proliferation via regulation of p21 and p57 expression levels31.
Recently, the ceRNA hypothesis proposed that lncRNAs communicate with other protein-coding RNA transcripts via shared common miRNA binding sites32. According to the ceRNA hypothesis, miRNA complementary base pairing with XIST was predicted by starBase and TargetScanS, and miR-449a was identified. Another reason is that our previous study showed that miR-449a suppressed cell invasion and promoted cell apoptosis in NSCLC18,33. However, quantitative real-time PCR showed that the expression of miR-449a was increased upon knockdown of XIST, and the expression of XIST was suppressed by miR-449a. To explore the molecular mechanism of how XIST influences cell growth in NSCLC, we performed site-directed mutagenesis of the XIST gene and used the luciferase reporter assay to detect the association between XIST and miR-449a. The results showed that XIST might be linked to miR-449a by the way of complementarity of the gene sequence. Western blot assay was performed in NSCLC cells that were co-transfected with shXIST and miR-449a inhibitor, and the results showed that the XIST-induced changes of Bcl-2 and PARP-1 protein were rescued. Furthermore, we upregulated the expression of XIST by transfection NSCLC cells with an miR-449a inhibitor. Wound healing and transwell assays were performed to detect the cell migration and invasion ability. The results demonstrated that the migratory and invasive abilities were enhanced when XIST was upregulated. On account of miRNA-449a promoting cell apoptosis by targeting Bcl-234, XIST can regulate the expression of Bcl-2 by competitively binding to miR-449a, which can regulate the NSCLC cell apoptosis.
In the present study, we revealed a previously uncharacterized role of XIST in regulating NSCLC cell growth. We also demonstrated that XIST functions as a ceRNA to attenuate the endogenous function of miR-449a. Both of these observed activities modulate Bcl-2 expression. Hence, by competing with Bcl-2 to bind miR-449a, XIST influences the regulation of tumor progression in NSCLC.
Author contribution
Ya-long ZHANG collected and analyzed data and wrote the manuscript. Xue-bing LI designed the study and wrote the manuscript. Yan-xu HOU and Nian-zhen FANG collected data. Jia-cong YOU designed the study and analyzed data. Qing-hua ZHOU designed the study.
Table 1. Primers sequence of quantitative real-time PCR.
| Gene | Sense primer (5′→3′) | Antisense primer (5′→3′) |
|---|---|---|
| U6 | TGCGGGTGCTCGCTTCGGCAGC | CCAGTGCAGGGTCCGAGGT |
| GAPDH | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA |
| miR-449a | TGCGGTGGCAGTGTATTGTTAGC | CCAGTGCAGGGTCCGAGGT |
| XIST | CTAGCTAGCTTTTGTAGTGAGCTTGCTCCT | TGCTCTAGAATGTCTCCATCTCCATTTTGC |
Acknowledgments
This study was supported by grants from the Ministry of Education Fund Priority to the Development of Instructions of Higher Leading Doctoral Degree Field (No 20131202130001, to Qing-hua ZHOU), the National Natural Science Foundation of China (No 81572288, to Qing-hua ZHOU; No 81302002, to Xue-bing LI), the Tianjin Natural Science Foundation (No 14JCQNJC12300, to Xue-bing LI) and the “New Century” Talent Training Project of Tianjin Medical University General Hospital (2014, to Xue-bing LI).
Footnotes
Supplementary information is available on the website of Acta Pharmacologica Sinica.
Supplementary Information
Sequences of siRNA for knockdown of XIST.
Sequences of shRNA embedded in lentivirus for knockdown of XIST.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- Goldstraw P, Ball D, Jett JR, Le CT, Lim E, Nicholson AG, et al. Non-small-cell lung cancer. Lancet 2011; 378: 1727–40. [DOI] [PubMed] [Google Scholar]
- Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, et al. Recent cancer survival in Europe: a 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol 2007; 8: 784–96. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature 2001; 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science 2005; 309: 1559–63. [DOI] [PubMed] [Google Scholar]
- An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 2008; 105: 716–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 2006; 16: 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33: 717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23: 1494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003; 22: 8031–41. [DOI] [PubMed] [Google Scholar]
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011; 10: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Kong R, Yin DD, Zhang EB, Xu TP, De W, et al. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol 2013; 30: 694. [DOI] [PubMed] [Google Scholar]
- Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol 2013; 49: 204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. Fifty years of X-inactivation research. Development 2011; 138: 5049–55. [DOI] [PubMed] [Google Scholar]
- Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg 2011; 35: 1751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett 2015; 359: 75–86. [DOI] [PubMed] [Google Scholar]
- You J, Zhang Y, Liu B, Li Y, Fang N, Zu L, et al. MicroRNA-449a inhibits cell growth in lung cancer and regulates long noncoding RNA nuclear enriched abundant transcript 1. Indian J Cancer 2014; 51 Suppl 3: e77–81. [DOI] [PubMed] [Google Scholar]
- Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett 2015; 361: 13–21. [DOI] [PubMed] [Google Scholar]
- Roth A, Diederichs S. Long noncoding RNAs in lung cancer. Curr Top Microbiol Immunol 2016; 394: 57–110. [DOI] [PubMed] [Google Scholar]
- Zequn N, Xuemei Z, Wei L, Zongjuan M, Yujie Z, Yanli H, et al. The role and potential mechanisms of LncRNA-TATDN1 on metastasis and invasion of non-small cell lung cancer. Oncotarget 2016; 7: 18219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther 2015; 14: 268–77. [DOI] [PubMed] [Google Scholar]
- Zhou C, Ye L, Jiang C, Bai J, Chi Y, Zhang H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol 2015; 36: 9179–88. [DOI] [PubMed] [Google Scholar]
- Guo F, Guo L, Li Y, Zhou Q, Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol 2015; 8: 15903–10. [PMC free article] [PubMed] [Google Scholar]
- Lv J, Qiu M, Xia W, Liu C, Xu Y, Wang J, et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res 2016; 35: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 2013; 152: 727–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Gao S, He W, Kou X, Zhao Y, Wang H, et al. Xist repression shows time-dependent effects on the reprogramming of female somatic cells to induced pluripotent stem cells. Stem Cells 2014; 32: 2642–56. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458: 223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chen X, Xu T, Xia R, Han L, Chen W, et al. MiR-326 regulates cell proliferation and migration in lung cancer by targeting phox2a and is regulated by HOTAIR. Am J Cancer Res 2016; 6: 173–86. [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia 2014; 16: 900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renhua G, Yue S, Shidai J, Jing F, Xiyi L. 165P: Long noncoding RNA LUCAT1 is associated with poor prognosis in human non-small cell lung cancer and affects cell proliferation via regulating p21 and p57 expression. J Thorac Oncol 2016; 11: S129. [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell 2011; 146: 353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zhang Y, Li Y, Fang N, Liu B, Zu L, et al. MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am J Cancer Res 2015; 5: 2730–44. [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhou J, Chen X, Yang B, Wang D, Yang P, et al. miRNA-449a is downregulated in osteosarcoma and promotes cell apoptosis by targeting BCL2. Tumour Biol 2015; 36: 8221–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of siRNA for knockdown of XIST.
Sequences of shRNA embedded in lentivirus for knockdown of XIST.