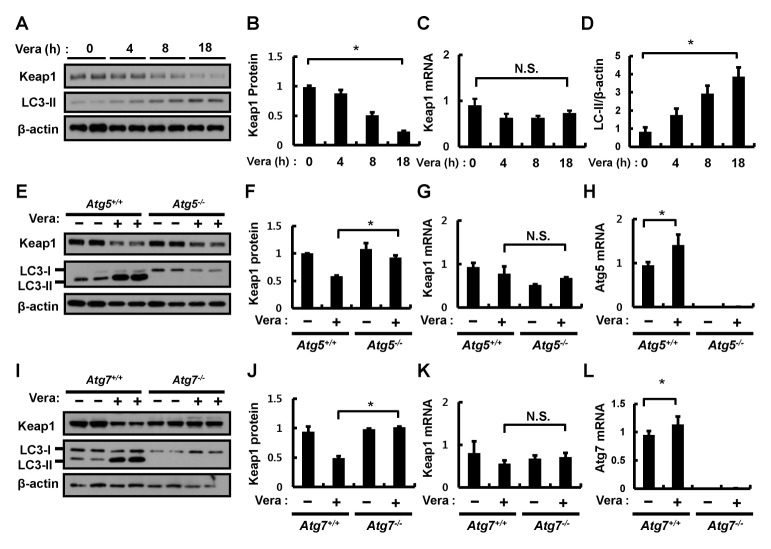
Fig. 2.
Verapamil mediates Keap1 degradation via an autophagic pathway. (A) Hepa1c1c7 cells were treated with verapamil (100 μM) for the indicated times. Lysates of Hepa1c1c7 cells were subjected to immunoblot analysis with antibodies against Keap1, LC3-II, and β-actin (loading control). (B) Densitometric analysis of Keap1 immunoblots was obtained in (A). Total RNA isolated from cells treated as described was subjected to qRT-PCR analysis for the mRNA of Keap1 (C). (D) Densitometric analysis of LC3-II/β-actin immunoblots was obtained in (A). (E) ATG5+/+ or ATG5−/− MEF cells were incubated in the absence or presence of verapamil (100 μM) for 18 h. Lysates of ATG5+/+ or ATG5−/− MEF cells were subjected to immunoblot analysis with antibodies against Keap1, LC3-II, and β-actin (loading control). (F) Densitometric analysis of Keap1 immunoblots was obtained in (E). Total RNA isolated from cells treated as described in (E) was subjected to qRT-PCR analysis for mRNAs of Keap1 (G) and ATG5 (H). (I) ATG7+/+ or ATG7−/− MEF cells were incubated in the absence or presence of verapamil (100 μM) for 18 h. Lysates of ATG7+/+ or ATG7−/− MEF cells were subjected to immunoblot analysis with antibodies against Keap1, LC3-II, and β-actin (loading control). (J) Densitometric analysis of Keap1 immunoblots was obtained in (H). Total RNA isolated from cells treated as described in (I) was subjected to qRT-PCR analysis for mRNAs of Keap1 (K) and ATG7 (L). Data are the mean ± SD from three independent experiments. *P < 0.05.