Figure 3. NMR structure determination of Pom152718–820 reveals a conserved Ig-like fold domain.
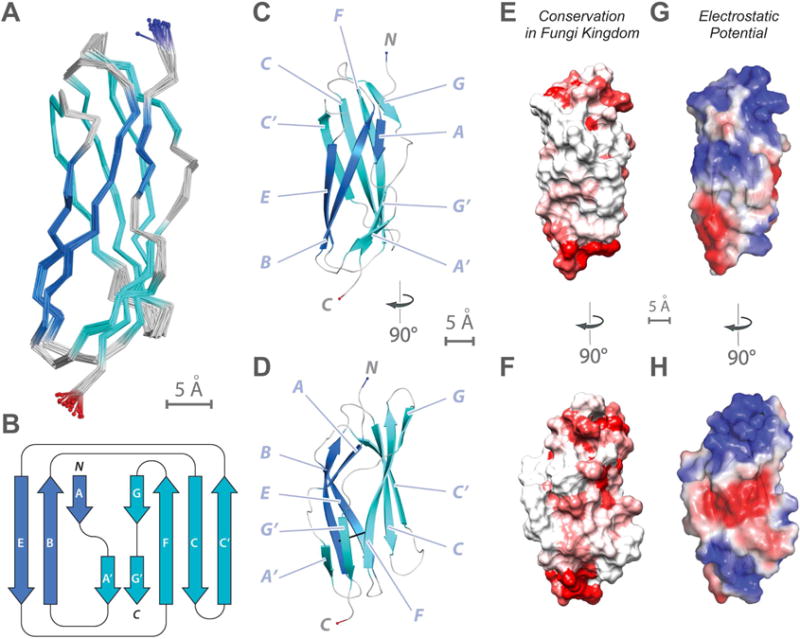
(A) Superimposition of 20 lowest-energy structures of Pom152718–820 that satisfy the NMR restraints best. The two β-sheets are made of the ABE (blue) and C′CFGG′A′ β-strands (cyan).
(B) Topology of the Pom152718–820 fold. β-strands are represented as thick arrows and the linkers connecting them as lines. Each strand is named following the standard Ig-like domain annotation (Halaby et al., 1999). Scale bar, 5 Å.
(C and D) Two views of the best-scoring structure, showing the arrangement of β-strands and features labeled as in panel B.
(E and F) Sequence conservation of Pom152718–820 among 37 fungal homologs (Figure S6), plotted on the surface of the Pom152718–820 structure; low conservation in white, high conservation in red.
(G and H) Electrostatic potential on the surface of the Pom152718–820 structure, calculated using the PyMOL tool APBS; negative (−2 kT/e) and positive (+2 kT/e) potentials are shown in red and blue, respectively.
