Figure 6. Integrative structure of Pom152FL based on NMR spectroscopy, negative-stain EM, and SAXS.
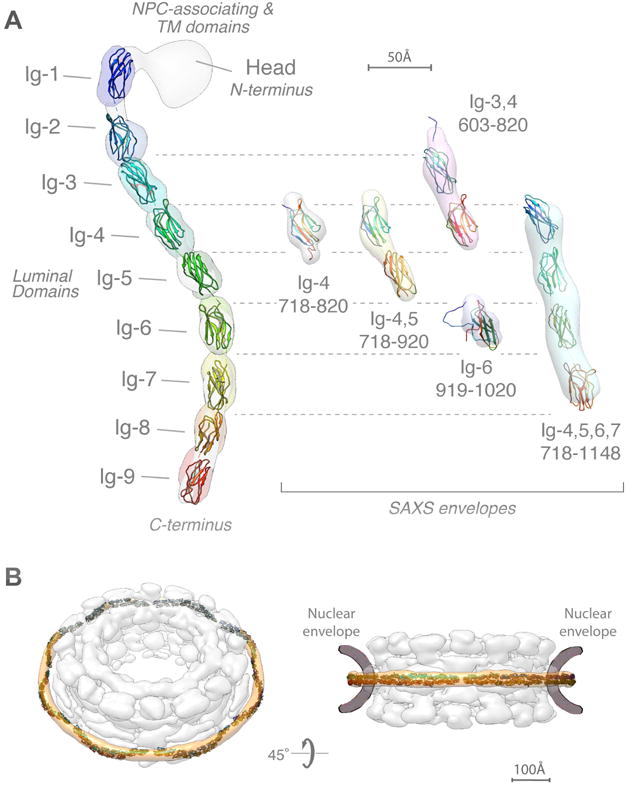
(A) (left) The localization probability density map computed from 364 superposed structures that satisfy the input spatial restraints shows the location of each of the 9 Ig-like domains (ranging from blue to red). The negative-stain EM density map is superposed in grey. A representative molecular model of Pom152LD (ribbon plot) was obtained by adjusting the relative orientations of adjacent Ig-like domains to resemble those in known cadherin structures (PDB codes 1L3W, 1EPF, 1NCI, 4ZI9, and 5K8R) (Boggon et al., 2002; Kasper et al., 2000; Nicoludis et al., 2015; Nicoludis et al., 2016; Shapiro et al., 1995).
(right) Validation of the integrative structure of Pom152LD by SAXS data for five Pom152 segments, spanning residues 718–820 (Ig-4), 718–920 (Ig-4,5), 603–820 (Ig-3,4), 919–1020 (Ig-6), and 718–1148 (Ig-4,5,6,7). The shapes of these segments in our integrative structure match the envelopes (ab initio shapes) computed from the corresponding SAXS profiles.
(B) Fit of 16 copies of Pom152LD into the yeast NPC map (Alber et al., 2007b). A good fit positions two copies of the extended Pom152LD molecule in an anti-parallel fashion on top of each other, forming a homodimer; we only show the potential arrangement of the antiparallel homodimer, which is implied by the C2-symmetry of the NPC (Alber et al., 2007b; Kosinski et al., 2016; Lin et al., 2016).
