Figure 2.
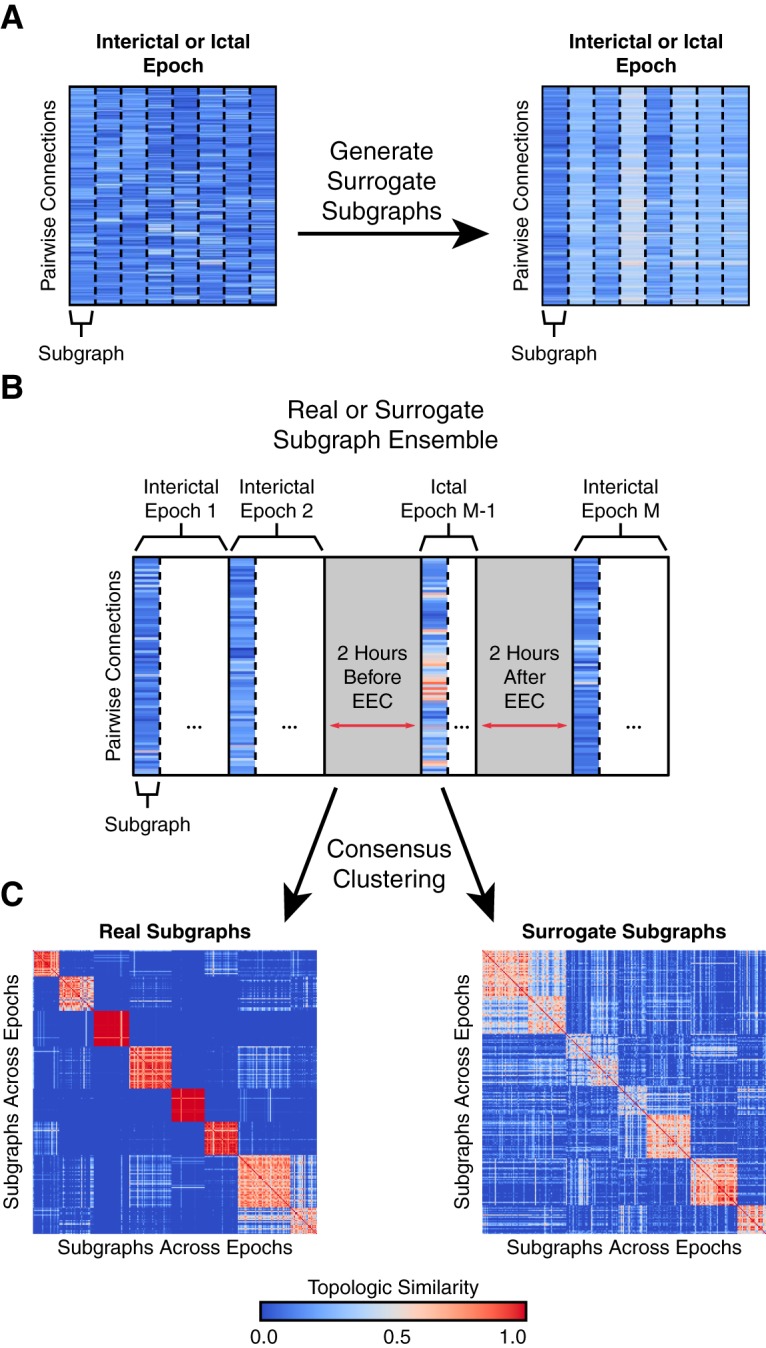
Clustering subgraphs based on topological similarity. A, For the set of original subgraphs learned from an epoch of data (left), we generated an equally sized set of surrogate subgraphs (right) by computing a weighted linear combination of the subgraphs with weights drawn from a uniform random distribution on the interval [0, 1]. The surrogate subgraphs have rewired network topology but maintain their functionality as a mathematical basis of the original network. B, For each patient, we constructed a subgraph ensemble matrix, representing the functional connections for each subgraph from all interictal and ictal epochs. The ensemble matrix aggregates functional subgraphs expressed over ∼100 h of intracranial recording. We also constructed a patient-specific surrogate ensemble matrix by aggregating surrogate subgraphs across all epochs. C, We quantified the topological similarity between all subgraphs in the ensemble matrix by applying a consensus NMF algorithm that tracks the number of times every pair of subgraphs is assigned to the same cluster over 100 runs of NMF. This procedure resulted in a coclustering probability matrix representing the frequency with which subgraphs from ictal and interictal epochs are clustered together, a measure of similarity between the connectivity profiles of subgraph pairs. In the example, the coclustering probability matrix of real subgraphs demonstrates less ambiguous similarity (matrix entries are near 0 or 1) and greater clustering than surrogate subgraphs (matrix entries closer to 0.5).
