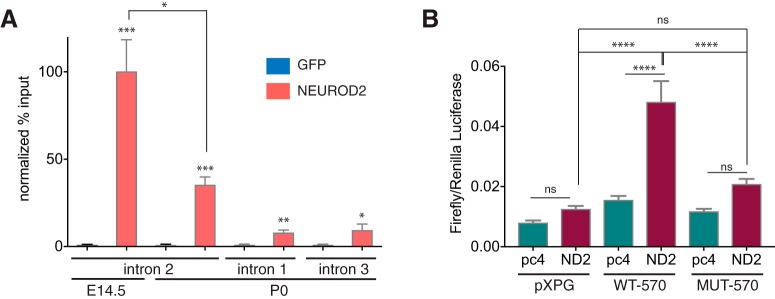
Figure 3.
Verification of NEUROD2 binding to the conserved element within Stim1 intron 2. A, NEUROD2 binding to Stim1 intronic element is confirmed in E14.5 and P0 cortices by ChIP-qPCR. ChIP DNA acquired with an unrelated GFP antibody is used as a negative control. Amount of DNA immunoprecipitated with either a NEUROD2 antibody (NEUROD2 ChIP DNA) or GFP antibody (GFP ChIP DNA) is expressed as percentage of input DNA (% input). NEUROD2 % input values are then normalized to GFP % input values. Strong enrichment of NEUROD2 is detected at the NEUROD2 binding element located in Stim1 intron 2 both in E14.5 and P0 cortices. Slight enrichments are observed for Stim1 introns 1 and 3. Data are representative of six biological replicates each composed of three technical replicates. Bars represent SEM. p < 0.0001 determined by one-way ANOVA followed by unpaired t test, *p < 0.05, **p < 1 × 10−4, ***p < 1 × 10−5 (Table 2). B, Luciferase activity is measured from HEK 293T cell lysates that are transfected either with an empty luciferase reporter plasmid (pXPG) or with a luciferase reporter downstream of a wild-type (WT-570) or mutated 570 bp fragment (MUT-570) Stim1 intronic element. In addition, cells are also cotransfected with either an empty (pc4) or NEUROD2 expressing (ND2) pcDNA4 vector. Firefly luciferase activity is normalized to Renilla luciferase signal. Data represent three independent experiments with each sample measured in triplicates. Bars represent SEM. D’Agostino–Pearson test showed normal distribution of the data (α = 0.05). One-way ANOVA and post hoc Tukey’s multiple-comparison analysis was performed, ****p < 0.0001 (Table 2).