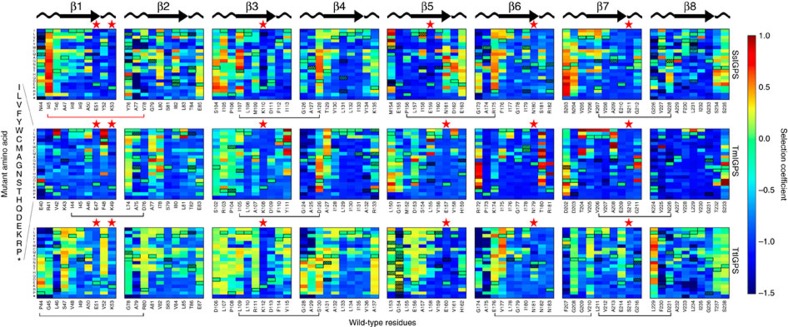
Figure 2. Fitness landscapes of the three IGPS orthologues.
Values of the selection coefficient are colour-coded on a continuous scale from 1 to −1.5 indicated by the colorbar. WT residues and positions are labelled at the bottom of each panel. Mutant amino acids are indicated on the vertical axis. Within the heatmap, WT residues are indicated by the black outline. Low-quality data filtered from analysis are indicated by the red checkered boxes. Canonical secondary structures are drawn at the top of the panels. Active sites are indicated by the red stars at the top of the position columns. βα hairpin clamps are indicated by the black brackets at the bottom of the position columns. Fitness gains were observed with several mutations of the βα hairpin clamps. For example, more than half the mutations in the three β3α3 hairpin clamps SsIGPS I107 and D128, TmIGPS I105 and D126, TtIGPS I109 and S130 are beneficial (s>0). Two non-canonical interactions analogous to the βα hairpin clamp in SsIGPS are indicated by the red brackets. An ionic interaction is observed between E155 and R175. A hydrophobic stacking interaction is observed between I45, V78, and F40 (not included in library). All mutations of SsIGPS I45, except to stop codons, resulted in fitness advantage over WT.