Abstract
During development, cortical interneurons generated from the ventral telencephalon migrate tangentially into the dorsal telencephalon. Although Achaete-scute family bHLH transcription factor 1 (Ascl1) plays important roles in the developing telencephalon, whether Ascl1 regulates tangential migration remains unclear. Here, we found that Ascl1 promoted tangential migration along the ventricular zone/subventricular zone (VZ/SVZ) and intermediate zone (IZ) of the dorsal telencephalon. Distal-less homeobox 2 (Dlx2) acted downstream of Ascl1 in promoting tangential migration along the VZ/SVZ but not IZ. We further identified Eph receptor B2 (Ephb2) as a direct target of Ascl1. Knockdown of EphB2 disrupted the separation of the VZ/SVZ and IZ migratory routes. Ephrin-A5, a ligand of EphB2, was sufficient to repel both Ascl1-expressing cells in vitro and tangentially migrating cortical interneurons in vivo. Together, our results demonstrate that Ascl1 induces expression of Dlx2 and Ephb2 to maintain distinct tangential migratory routes in the dorsal telencephalon.
The cerebral cortex is important for executing high-order brain functions, such as sensory perception, motor control and cognition. Proper functions of the cerebral cortex require coordination between excitatory (glutamatergic) projection neurons and inhibitory (GABAergic) interneurons. Many neurological and psychological disorders, such as epilepsy, autism, schizophrenia, and Alzheimer’s disease, may be resulted from an imbalance of excitatory and inhibitory neuronal activities or the dysfunction of interneurons1,2. Interestingly, glutamatergic neurons and GABAergic interneurons are produced from different regions of the developing brain. Glutamatergic neurons are generated from neural progenitors in the dorsal telencephalon, which develops into the cerebral cortex. GABAergic cortical interneurons are remotely derived from neural progenitors in the ventral telencephalon, which develops into the striatum and globus pallidus3. Around 85% of cortical interneurons are generated from the medial ganglionic eminence (MGE) and the rest are generated from the caudal ganglionic eminence (CGE)4,5.
Once generated in the ventricular zone (VZ), glutamatergic neurons and GABAergic interneurons migrate toward final destinations through distinct paths. Glutamatergic neurons undergo radial migration to reach the cortical plate (CP)6. Interneurons take a long tangentially migratory path from the ventral telencephalon toward the striatum or the cerebral cortex3. When cortical interneurons reach the dorsal telencephalon, they continue migrating tangentially through three migratory routes: the marginal zone (MZ), intermediate zone (IZ) or ventricular/subventricular zone (VZ/SVZ) of the dorsal telencephalon7,8. Upon arrival at destined region, interneurons switch to migrate radially until they reach specific layers of the cerebral cortex, where they integrate into local neuronal circuits. Since cortical interneurons go through such a complicated migratory path, it is important to study the underlying regulatory mechanism.
Several transcription factors control cell fate specification, differentiation and migration in the telencephalon. Paired Box 6 (Pax6) and Neurogenin 1 and 2 (Neurog1/2) are expressed in the dorsal telencephalon and regulate cell fate specification and differentiation of glutamatergic neurons9,10. In addition, Neurog2 and Ascl1 induce expression of small GTPase genes to modulate actin dynamics therefore controlling radial migration of glutamatergic neurons11,12. In the ventral telencephalon, Dlx1 and 2, NK2 homeobox 1 (Nkx2.1), and Ascl1 specify and promote differentiation of GABAergic interneurons13. In addition, Dlx1/2 and Nkx2.1 orchestrate regulators for cytoskeleton dynamics, mitogenic cues and chemotactic molecules to control tangential migration14,15. While Ascl1 regulates radial migration of glutamatergic neurons in the dorsal telencephalon11,16, Ascl1 is also abundantly expressed in the ventral telencephalon and critical for specification and differentiation of neurons generated from the ventral telencephalon10,17. However, whether Ascl1 induces genes involved in regulating tangential migration remains unknown.
Previous studies have suggested that Ascl1 might play roles in tangential migration. Nearly 60% of the Ascl1-expressing neurons migrate tangentially along the VZ/SVZ and IZ routes in the dorsal telencephalon18. Mutation of Ascl1 leads to reduction of cortical interneurons in mice19, suggesting that Ascl1 is required for specification and possibly tangential migration of cortical interneurons. Furthermore, Ascl1 induces the expression of Dlx1/2 in the ventral telencephalon17,20. Since Dlx1/2 regulate the expression of genes involved in tangential migration15,21,22, it is possible that Ascl1 acts upstream of Dlx1/2 to regulate tangential migration of cortical interneurons.
Here, we used in utero electroporation to investigate the role of Ascl1 in tangential migration. We found that Ascl1 promoted tangential migration through both VZ/SVZ and IZ routes in the dorsal telencephalon, and Dlx2 acted downstream of Ascl1 to promote tangential migration through the VZ/SVZ route. Furthermore, Ascl1 induced Ephb2, which mediated the separation of the VZ/SVZ and IZ migratory routes. Ephrin-A5, a binding partner of EphB2, had a repulsive effect on Ascl1-expressing neurons in vitro and cortical interneurons in vivo. Together, our work provides evidence that Ascl1 induces Ephb2 and the interaction between EphB2 and Ephrin-A5 is crucial for restricting interneurons to migrate on distinct routes in the dorsal telencephalon.
Results
Ascl1 and Dlx2 regulate the migratory behavior in the ventral telencephalon
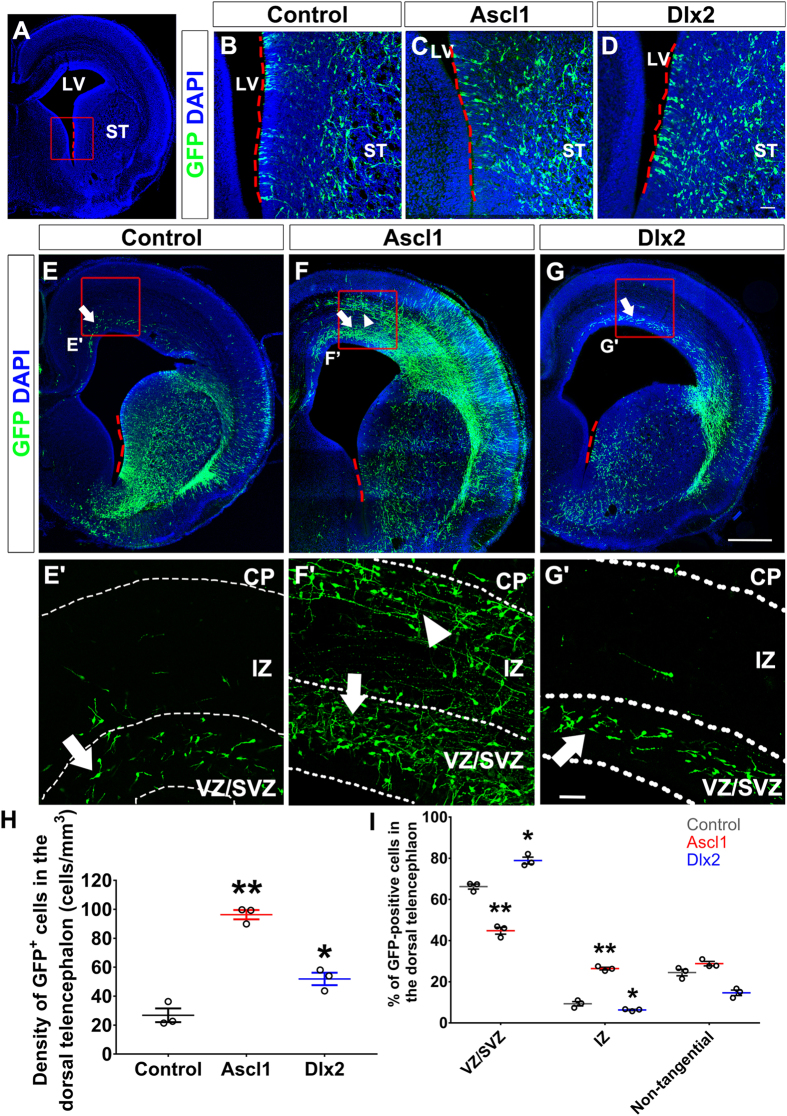
Although knockout of Ascl1 or Dlx1/2 reduces the number of interneurons reaching the dorsal telencephalon in mice19,23, whether Ascl1 or Dlx2 promotes tangential migration has never been tested. Thus, we transfected Ascl1 or Dlx2 together with GFP expression constructs into the ventral telencephalon of rats at embryonic day 15.5 (E15.5) by using in utero electroporation (Fig. 1A). The parental vector US2 was used as a control. Four days after electroporation, many GFP-positive cells were observed near the ventricle of the striatum (Fig. 1B–D), demonstrating that these constructs were delivered into the ventral telencephalon successfully. In the control group, some GFP-positive cells were present in the VZ/SVZ of the dorsal telencephalon with leading processes in parallel with the ventricle (Fig. 1E), suggesting that these neurons had migrated from the ventral to the dorsal telencephalon and were migrating along the VZ/SVZ route. Overexpression of Ascl1 or Dlx2 dramatically increased the number of GFP-positive cells in the dorsal telencephalon (Fig. 1F–H; the density of GFP-positive cells in the dorsal telencephalon is 26.8 ± 4.8 cells/mm3 in control, 96.3 ± 3.2 cells/mm3 in Ascl1 group, and 51.9 ± 4.3 cells/mm3 in Dlx2 group, n = 3). Interestingly, overexpression of Ascl1 increased GFP-positive cells in both the VZ/SVZ and IZ, while overexpression of Dlx2 only increased GFP-positive cells in the VZ/SVZ of the dorsal telencephalon (Fig. 1F,G). Quantitative analysis further demonstrated that overexpression of Ascl1 increased the percentage of GFP-positive cells in the IZ, while overexpression of Dlx2 increased the percentage of GFP-positive cells in the VZ/SVZ of the dorsal telencephalon (Fig. 1I). Microtubule-associated protein 2 (MAP2) is detected in differentiating and differentiated neurons. Overexpression of Ascl1 did not increase the percentage of GFP- and MAP2-double positive cells in the dorsal telencephalon four days after electroporation (Fig. S1), which ruled out the possibility that overexpression of Ascl1 increased GFP-positive cells in the dorsal telencephalon through promoting neuronal differentiation. Thus, this result suggests that Ascl1 promotes tangential migration through both VZ/SVZ and IZ routes, while Dlx2 promotes tangential migration only through the VZ/SVZ route.
Figure 1. Overexpression of Ascl1 and Dlx2 in the ventral telencephalon promotes tangential migration.
Four days after electroporation, brains of E19.5 rats were dissected and sectioned in the coronal plane. Electroporated cells were labeled with anti-GFP in green; nuclear DNA was stained with DAPI in blue. Red dashed lines indicate the electroporation region in (A–G). (A) A confocal image of brain section; red dashed lines mark the electroporated region and a red square indicate the area for (B–D). ST: striatum, LV: lateral ventricle. (B–D) GFP-positive cells were distributed in the ST adjacent to the LV. (E) In the control group, some GFP-positive cells were distributed in the VZ/SVZ (white arrows) of the dorsal telencephalon, while most remained in the ventral telencephalon. (F) In Ascl1 group, GFP-positive cells were distributed in both the VZ/SVZ (white arrows) and IZ (white arrowheads) of the dorsal telencephalon. (G) In Dlx2 group, many GFP-positive cells were distributed in the VZ/SVZ (white arrow). Red squares indicate the zoom-in areas for 1E’ to G’. Length of the scale bar is 40 μm in (B–D) and (E’–G’), 250 μm in (E–G). (H) Quantification of GFP-positive cell density in the dorsal telencephalon. (I) Quantification of GFP-positive cells in the VZ/SVZ, IZ, or non-tangentially migrating cells in the dorsal telencephalon. Data were presented as mean ± standard error of the mean (SEM) with all data points and analyzed by Student’s t-test, n = 3. *p < 0.05; **p < 0.01 compared to the control group.
Ascl1 and Dlx2 are sufficient to promote tangential migration in the dorsal telencephalon
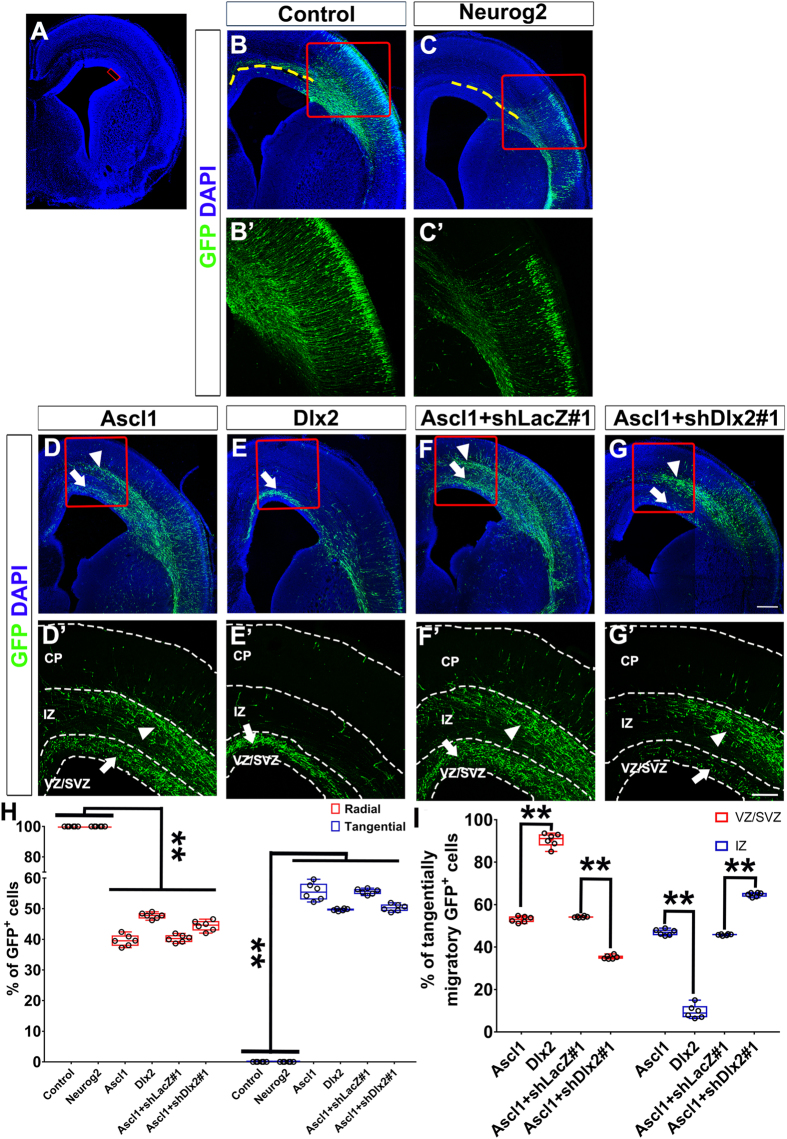
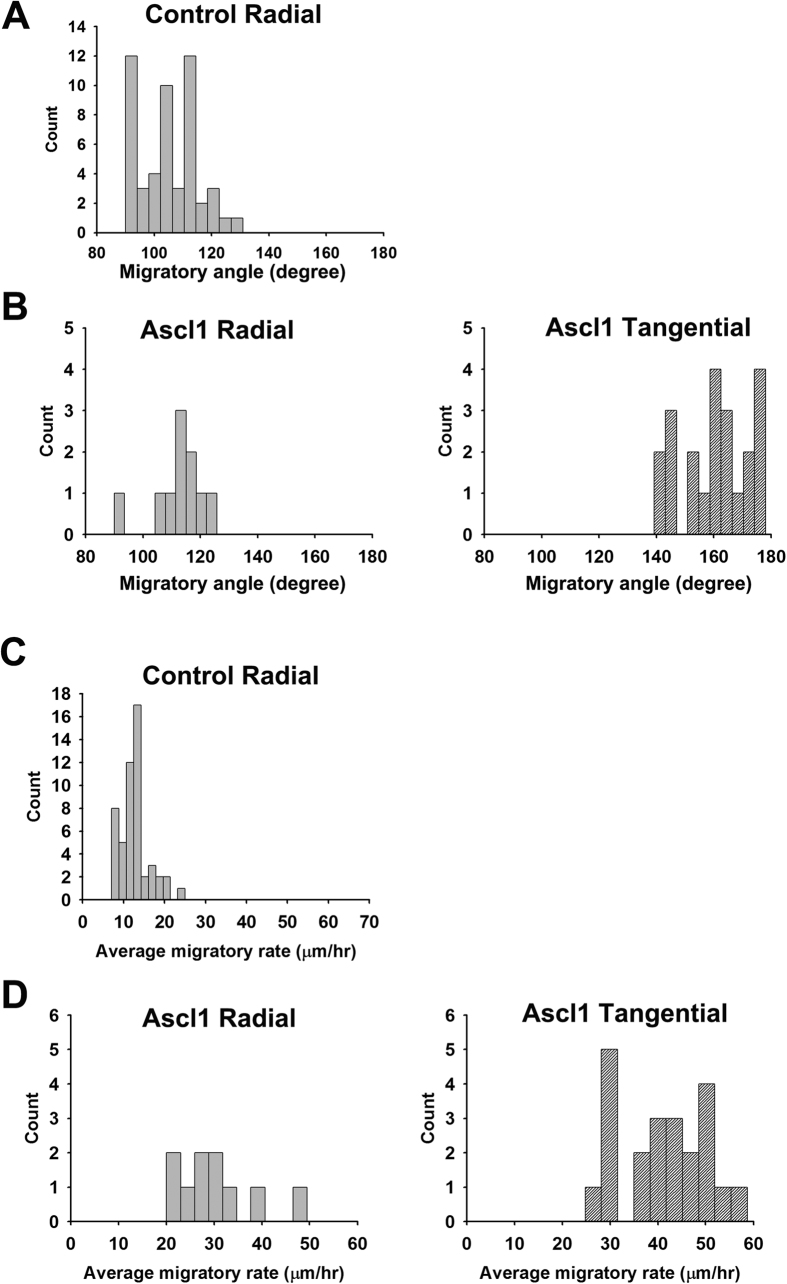
While cortical interneurons migrate tangentially into the dorsal telencephalon, glutamatergic neurons are generated in the dorsal telencephalon and migrate radially toward the CP. Thus, it would be intriguing to test whether radial migration program in neurons generated from the dorsal telencephalon can be switched to tangential migration program driven by Ascl1 or Dlx2. Expression constructs for Ascl1 , Dlx2 or Neurog2 was electroporated into the dorsal telencephalon together with a GFP expression construct (Fig. 2A). In the control group, some GFP-positive cells were distributed near the electroporated region and many GFP-positive cells reached the CP four days after electroporation (Fig. 2B). Most of these cells extended their leading processes toward the CP, suggesting that they were migrating radially (Fig. 2B’). Overexpression of Neurog2 decreased the proportion of GFP-positive cells in the VZ/SVZ and increased the proportion of GFP-positive cells in the CP (Fig. 2C), confirming that Neurog2 promotes differentiation and radial migration in the dorsal telencephalon12. Similar to our findings in the ventral telencephalon, overexpression of Ascl1 or Dlx2 in the dorsal telencephalon increased GFP-positive cells dorsomedial to the electroporation site (Fig. 2D,E). Many GFP-positive cells in the VZ/SVZ and IZ extended their processes in parallel with the ventricle, suggesting that they were migrating tangentially (Fig. 2D’,E’). We categorized those GFP-positive cells into three groups according to the orientation of their leading processes: cells with a horizontal leading process were categorized as tangentially migrating cells; cells with a vertical leading process were radially migrating cells; cells without a distinguishable leading process were “others” (Fig. S2). Quantitative analysis showed that overexpression of Ascl1 or Dlx2 increased the percentage of tangentially migrating cells (Fig. 2H; the percentage was 0% in the control group, 55.3 ± 1.6% in Ascl1 group, and 49.7 ± 1.4% in Dlx2 group, n = 6). Overexpression of Ascl1 increased the percentages of tangentially migrating GFP-positive cells in both IZ and VZ/SVZ; overexpression of Dlx2 only increased the percentage of tangentially migrating GFP-positive cells in the VZ/SVZ (Fig. 2D,E,I; the percentage of GFP-positive cells in VZ/SVZ and IZ were 55.2 ± 1.2% and 46.8 ± 2.8% in Ascl1 group, 85.0 ± 2.0% and 15.0 ± 2.3% in Dlx2 group, n = 6). Consistently, overexpression of Ascl1 or Dlx2 in the dorsal telencephalon resulted in a similar distribution pattern of GFP-positive cells in mice as that in rats (Fig. S3). We further recorded live imaging of brain slices from control and Ascl1s group two days after electroporation (Fig. 3 and Supplementary videos). In the control group, most GFP-positive cells migrated radially (Fig. 3A, Video S1). In Ascl1 group, GFP-positive cells migrated either radially or tangentially (Fig. 3B, Video S2). Interestingly, the average migratory rate was dramatically increased in Ascl1 group in comparison with the control group (Fig. 3C,D), suggesting that Ascl1 promotes motility. No MAP2-positive cell was detected two days after electroporation in the IZ and VZ/SVZ of the dorsal telencephalon of control or Ascl1 group (Fig. S4), which ruled out the possibility that overexpression of Ascl1 changed GFP-positive cell distribution through accelerating neuronal differentiation.
Figure 2. Overexpression of Ascl1 in the dorsal telencephalon induces ectopically tangential migration.
Four days after electroporation to the dorsal telencephalon, brains of E19.5 rats were dissected and sectioned in the coronal plane. Electroporated cells were labeled with anti-GFP in green; nuclear DNA was stained with DAPI in blue. (A) A confocal image with a red square indicating the electroporated site. (B’–G’) are zoomed regions of (B–G) indicated by red squares. (B) In the control group. GFP-positive cells were distributed near the electroporated area and many of them extended processes radially. GFP-positive axons toward the contralateral side were observed (yellow dashed lines). (C) In Neurog2 group, most GFP-positive cells were distributed in the cortical plate (CP). GFP-positive axons toward the contralateral side were observed (yellow dashed lines). (D) In Ascl1 group, many GFP-positive cells were distributed in the VZ/SVZ (white arrows) and IZ (white arrowheads) dorsomedially to the electroporated site. (E) In Dlx2 group, many GFP-positive cells were distributed in the VZ/SVZ (white arrows) dorsomedially to the electroporated site. (F) In Ascl1 + shLacZ#1 group, GFP-positive cells were distributed in a similar pattern as Ascl1 group in (D). (G) In Ascl1+shDlx2#1 group, many GFP-positive cells were distributed in the IZ (white arrowheads) dorsomedially to the electroporated site. Few GFP-positive cells were distributed in the VZ/SVZ (white arrows). Length of the scale bar is 120 μm in (B–G), and 100 μm in (B’ to G’). (H) GFP-positive cells were categorized into radial, tangential, or other types according to the orientation of their leading processes. (I) Quantification of GFP-positive cells in the VZ/SVZ and IZ. Data are presented as box and whisker plots with all data points. The horizontal line within the box indicates the median, boundaries of the box indicate the 25th and 75th percentiles, and the whiskers indicate the maximum and minimum values of the results. Data are analyzed by using Student’s t-test, n = 6 in all groups. *p < 0.05; **p < 0.01.
Figure 3. Overexpression of Ascl1 changes the migratory behavior of neurons in the dorsal telencephalon.
Ascl1 or US2 control expression construct were co-electroporated with a GFP expression plasmid. Two days after electroporation to the dorsal telencephalon, brains of E17.5 rats were dissected and sectioned in the coronal plane for slice culture and live imaging recording. The video from 5 to 11 hours after recording was used for tracking the migratory behavior. We set 0° to 180° axis in parallel to the lateral ventricle and the dorsomedial side as 180°. The line connecting the cell body location in the first frame (starting point) and the last frame (end point) of the video was used for measuring migratory angle. The average migratory rate was calculated as accumulated distance of every six-minute interval divided by recording time. 50 GFP-positive cells were counted for the control and 32 were counted for Ascl1 group. All counts were plotted into histograms according to their migratory angle or rate. (A) In the control group, most GFP-positive cells migrated radially in a migratory angle between 90° to 130°. (B) In Ascl1 group, GFP-positive cells were categorized into radially (90°–130°) and tangentially (140°–180°) migrating cells. (C) In the control group, GFP-positive cells migrated in the rate of 12.9 ± 3.6 μm/hour (mean ± SEM). (D) In Ascl1 group, radially migrating cells migrated in the rate of 30.2 ± 8.7 μm/hour and tangentially migrating cells migrated in the rate of 40.8 ± 9.3 μm/hour.
To examine the possibility that Ascl1 or Dlx2 promotes tangential migration through specification of GABAergic neuronal fate, we examined markers of glutamatergic and GABAergic neurons. While most GFP-positive cells were positive for a pan-neuronal marker NeuN (96.7 ± 1.8%, n = 2) at postnatal day 13, they were not positive for the intermediate progenitor cell marker Eomesodermin (Tbr2) in the dorsal telencephalon, the glutamatergic neuronal marker vesicular Glutamate transporter 2 (vGlut2) or a GABAergic neuronal marker Gad67 at E19.5 and postnatal day 13 in rats (data not shown). Because Gad67 was difficult to detect by using immunofluorescence during embryonic stage, we used Gad67-GFP mice, whose GABAergic interneurons were labeled by Gad67 endogenous promoter-driven GFP24. Control, Ascl1- or Dlx2-expression constructs were electroporated with a DsRed-expression construct into the dorsal telencephalon of E14.5 Gad67-GFP mouse embryos. Four days after electroporation, no DsRed and GFP double-positive cells were identified in the control group (Fig. S5A,D), showing that the dorsal progenitors do not differentiate into GABAergic neurons. 13.6 ± 0.8% of Ascl1-overexpressing cells and 36.0 ± 3.7% of Dlx2-overexpressing cells were Gad67-GFP-positive (Fig. S5B–D). In these DsRed and GFP double positive cells, 35.2 ± 4.2% of Ascl1-overexpressing cells and 91.0 ± 2.8% of Dlx2-overexpressing cells were categorized as tangentially migrating neurons (Fig. S5B,C and E). We have previously shown that over 45% of Ascl1- or Dlx2-expressing cells underwent tangential migration four days after electroporation in the dorsal telencephalon in rats (Fig. 2H). If Ascl1 and Dlx2 promote tangential migration secondary to GABAergic neuronal fate specification, we expect to see higher percentages of Ascl1- and Dlx2-overexpressing cells adapt GABAergic fate and positive for Gad67-GFP. Thus, our data suggest that Ascl1 and Dlx2 might promote tangential migration in parallel with re-specification of GABAergic neuronal fate.
Ascl1 promotes tangential migration through Dlx2-dependent and Dlx2-independent manners
It has been shown that Dlx2 is a direct target gene of Ascl120. Since both Ascl1 and Dlx2 promote tangential migration, it is possible that Ascl1 promotes tangential migration by inducing Dlx2. To test it, expression constructs for Ascl1 and shRNAs targeting Dlx2 (shDlx2#1 and #2) were co-electroporated into the dorsal telencephalon. shLacZ#1 and #2 targeting LacZ were used as controls (Fig. S6). Transfection of shDlx2#1 or #2 efficiently knocked down Dlx2 in P19 cells, a mouse teratocarcinoma cell line (Fig. S6B). The distribution of GFP-positive cells in the dorsal telencephalon expressing Ascl1 and shLacZ#1 or #2 was similar to that of expressing Ascl1 alone (Figs 2D,F, S7B). Overexpression of Ascl1 and knockdown of Dlx2 did not reduce the percentage of tangentially migrating cells (Fig. 2F–H; the percentage was 55.7 ± 1.3% in Ascl1+shLacZ#1 group, 50.5 ± 1.6% in Ascl1+shDlx2#1 group, n = 6). Interestingly, the percentage of GFP-positive cells in the VZ/SVZ was reduced (Fig. 2F,G,I; the percentage of Ascl1 + shLacZ#1 group was 53.9 ± 0.9%, Ascl1+shDlx2#1 group was 36.8 ± 0.9%, n = 6), while the percentage of GFP-positive cells in the IZ was increased (Fig. 2F,G,I; the percentage of Ascl1 + shLacZ#1 group was 46.3 ± 0.4%, Ascl1+shDlx2#1 group was 63.2 ± 1.1%, n = 6). A similar distribution of GFP-positive cells was observed when we knocked down Dlx2 with shDlx2#2 (Fig. S7C). This result suggests that Dlx2 acts downstream of Ascl1 in promoting tangential migration through the VZ/SVZ route. In addition, Ascl1 promotes tangential migration through the IZ route independently of Dlx2.
EphB2 acts downstream of Ascl1 in regulating tangential migration
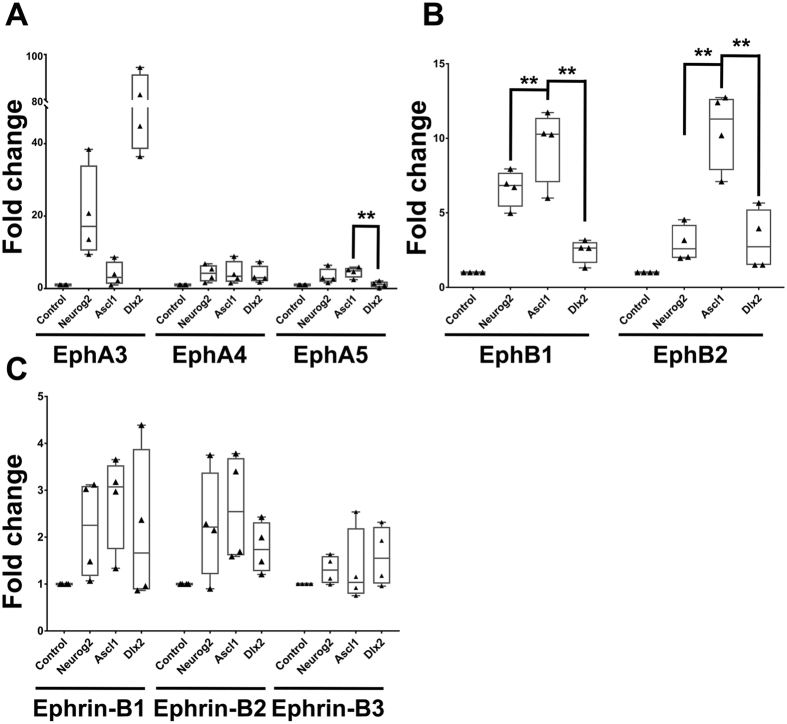
Although Rnd3 is shown to act downstream of Ascl1 to promote radial migration of glutamatergic neurons11, direct targets of Ascl1 involved in tangential migration of cortical interneurons have never been identified. Since overexpression of Ascl1 led to a distinct cell distribution in the IZ and VZ/SVZ (Figs 1 and 2), it is possible that Ascl1-expressing cells sense repulsive cues and avoid specific areas of the dorsal telencephalon. Eph and Ephrin families are repulsive cues for axons and migrating neurons25,26. Therefore, we examined whether Ascl1 induced expression of Ephs or Ephrins (Efns). P19 cells differentiate into neurons upon overexpression of Ascl1 or Neurog227,28, so we used P19 cells as a model to identify putative target genes of Ascl1. Expression constructs for Neurog2 , Ascl1, or Dlx2 were transfected into P19 cells and total RNA was extracted two days after transfection. By using quantitative RT-PCR (qRT-PCR), we examined the expression level of Ephs and Efns that have been detected in the developing telencephalon29. Because Neurog2 promotes differentiation of glutamatergic neurons17, genes induced by both Neurog2 and Ascl1 were excluded. Since Ascl1 activates Dlx217,20, genes induced by Dlx2 preferentially may be indirectly regulated by Ascl1. Thus, genes induced at higher levels by Ascl1 than Neurog2 and Dlx2 were selected as Ascl1 targets. Ephb1 and b2 mRNA were induced at higher levels by expression of Ascl1 than that of control and expression of Neurog2 or Dlx2 48 hours after transfection (Fig. 4). Ephb1 and b2 were induced dramatically: 9.2 ± 1.2 fold and 9.4 ± 1.1 fold relative to the control, respectively. Thus, we selected Ephb1 and b2 as candidate target genes and tested whether they act downstream of Ascl1 in regulating tangential migration.
Figure 4. Identification of Ascl1 target genes of the Eph and Ephrin (Efn) family.
P19 cells were transfected with Neurog2 , Ascl1, or Dlx2 expression constructs and total RNA was extracted two days after transfection. The expression of Ephs (A,B) and Efns (C) were normalized to the expression of TATA-box binding protein (Tbp). Ephb1 and Ephb2 were expressed at higher levels in Ascl1-expressing cells than those in Neurog2 and Dlx2-expressing cells. Data are presented as box and whisker plots with all data points. Data are analyzed by using Student’s t-test, n = 4. *p < 0.05; **p < 0.01.
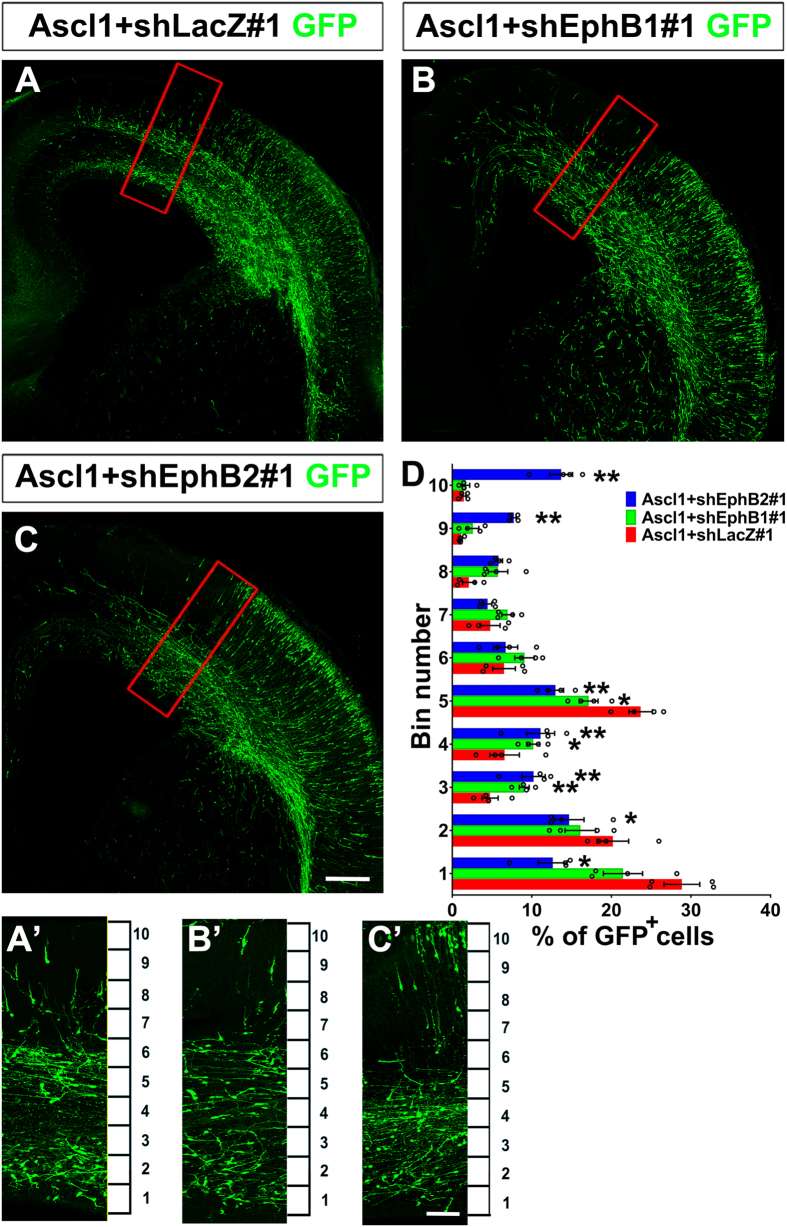
Expression constructs for Ascl1 and shRNAs against Ephb1 or b2 were electroporated into the dorsal telencephalon. Ephb1 or b2 mRNA expression was effectively inhibited by shRNAs against each gene (Fig. S6B). Knockdown of Ephb1 or b2 in Ascl1-overexpressing cells affected the distribution of GFP-positive cells. Importantly, the separation between the VZ/SVZ and IZ routes was disrupted (Fig. 5A–C, Fig. S7D,E). For quantification, the developing cortex was divided into 10 bins and the number of GFP-positive cells in each bin was counted. In Ascl1 + shLacZ#1 group, many GFP-positive cells were in bin 1 and 2 (the VZ/SVZ), as well as bin 5 (the IZ) (Fig. 5A’,D). Expression of Ascl1 and knockdown of Ephb1 slightly decreased the percentage of GFP-positive cells in bin 1, 2, and 5, but increased the percentage of cells in bin 3 and 4 comparing with Ascl1 + shLacZ#1 (Fig. 5B’,D). Expression of Ascl1 and knockdown of Ephb2 decreased the percentage of GFP-positive cells in bin 1, 2, and 5, but increased the percentage of cells in bin 3, 4, 9, and 10 comparing with Ascl1 + shLacZ#1 (Fig. 5C’,D). Knockdown of Ephb1 or b2 alone in the dorsal telencephalon did not affect distribution of electroporated cells in the CP (Fig. S8), showing that Ephb1 and b2 are not required for radial migration. These data suggest that EphB1 and B2 act downstream of Ascl1 in regulating the separation of the VZ/SVZ versus IZ routes during tangential migration.
Figure 5. Knockdown of Ephb1 or Ephb2 disrupted tangential migration promoted by Ascl1.
Four days after electroporation to the dorsal telencephalon, brains of E19.5 rats were dissected and sectioned in the coronal plane. Electroporated cells were labeled with anti-GFP in green. (A’–C’) are zoomed regions of (A–C) indicated by red squares, which were dorsomedial to the electroporated site. The cortex was divided equally into 10 bins. (A) In Ascl1 + shLacZ#1 group, many GFP positive cells were distributed in the VZ/SVZ (bins 1 and 2) and IZ (bin 5). (B,C) In Ascl1 + shEphB1#1 and Ascl1 + shEphB2#1 groups, GFP-positive cells were distributed in the VZ/SVZ (bins 1 and 2) and IZ (bin 5) were reduced. Length of the scale bar is 120 μm in (A–C), and 40 μm in (A’–C’). (D) Distribution of GFP-positive in the cortex. Only GFP-positive cells dorsomedial to the electroporated site were counted. Data were presented as mean ± SEM with all data points and analyzed by one-way ANOVA with Tukey’s-HSD post hoc test, n = 4. *p < 0.05; **p < 0.01 compared to Ascl1 + shLacZ#1 group.
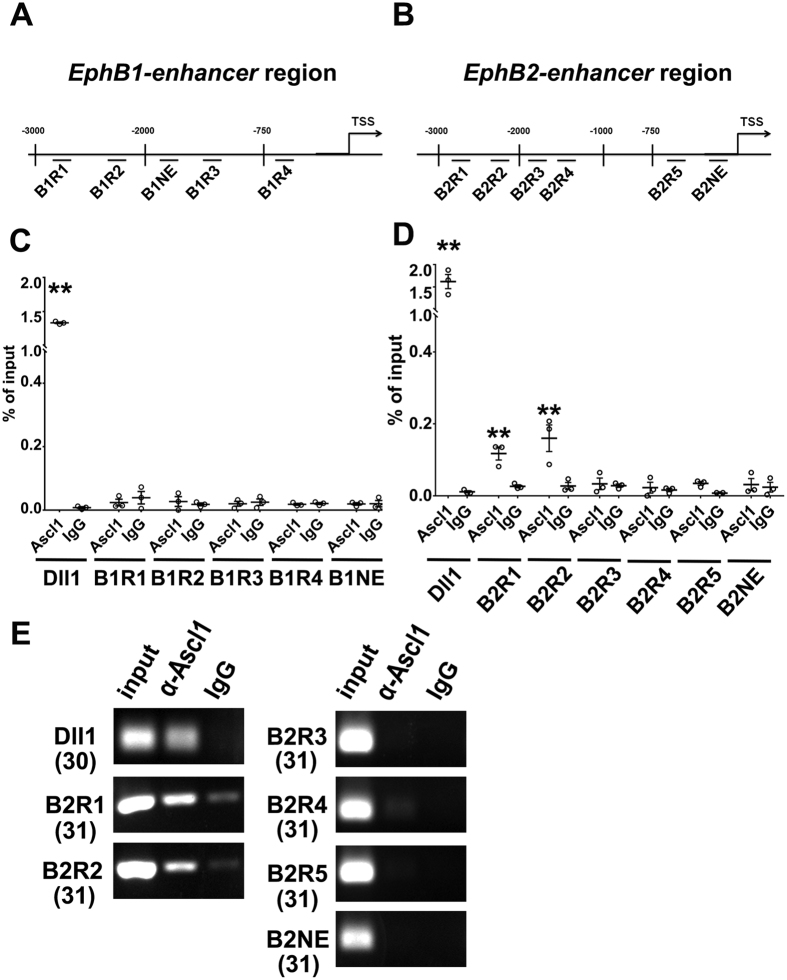
We further used Chromatin immunoprecipitation (ChIP) to check whether Ephb1 and b2 were direct targets of Ascl1. We analyzed three kilo-bases (Kb) upstream of the transcription start site (TSS) for E-box sequences (CACCTG, CAGATG, or CAGGTG) that are potential binding sites for Ascl130,31. Four putative E-boxes upstream of Ephb1 (designated as B1R1 to B1R4), and five upstream of Ephb2 (designated as B2R1 to B2R5) were identified (Fig. 6A,B). DNA was extracted from P19 cells transfected with an Ascl1 expression construct. A DNA fragment of Delta-like 1 (Dll1) with confirmed E-boxes was used as a positive control32. B1NE upstream of Ephb1 and B2NE upstream of Ephb2 without putative E-box were selected as negative controls. None of the DNA fragments upstream of Ephb1 was precipitated by anti-Ascl1 (Fig. 6C), suggesting that Ascl1 may not induce Ephb1 directly. Alternatively, Ascl1 may interact with an Ephb1 enhancer outside of the 3Kb-fragment upstream of the TSS that we examined. B2R1 and B2R2 of Ephb2 were precipitated by anti-Ascl1 (Fig. 6D,E), demonstrating that Ascl1 binds Ephb2 directly. Neither the negative controls (B1NE and B2NE) nor B2R3, B2R4, B2R5 were precipitated by anti-Ascl1 (Fig. 6D,E). Thus, we identified Ephb2 as a direct target of Ascl1.
Figure 6. Ephb2 is a direct target of Ascl1.
(A,B) Putative E-box sequences were identified in the 3 Kb upstream of the transcription starting site (TSS) for Ephb1 and Ephb2. Four fragments (B1R1 to B1R4) of Ephb1 and five fragments (B2R1 to B2R5) of Ephb2 containing E-box sequences were designed for ChIP. Two fragments (B1NE and B2NE) without E-box sequences were selected as negative controls. (C,D) DNA from P19 cells transfected with Ascl1 expression vectors was extracted for ChIP. A fragment in the promoter region of Dll1 that has been demonstrated to interact with Ascl1 was used as a positive control. After ChIP, enrichment of the DNA fragments was quantified by qPCR (C,D) and regular PCR (E). (C) The promoter region of Dll1 was enriched by ChIP with anti-Ascl1. No fragment in Ephb1 was enriched. (D) The promoter region of Dll1, as well as B2R1 and B2R2 sites of Ephb2 were enriched by ChIP with anti-Ascl1. (E) Regular PCR result. The number in the parentheses indicated PCR cycle. Data were presented as mean ± SEM with all data points and analyzed by Student’s t-test, n = 3; *p < 0.05 compared to the IgG control.
We further examined whether EphB2 could be detected in tangentially migrating cortical interneurons. Overexpression of Ascl1 in P19 cells increased the signal intensity of EphB2 detected by immunofluorescent staining (Fig. S9), which is consistent with our qPCR data (Fig. 4). Knockdown of Ephb2 decreased the signal intensity of EphB2 (Fig. S9), demonstrating the specificity of anti-EphB2. Furthermore, in Gad67-GFP E18.5 mouse embryos, most GFP-positive cells in the VZ/SVZ of the dorsal telencephalon were also EphB2-positive (Fig. S10), confirming that EphB2 is expressed in tangentially migrating cortical interneurons.
Ephrin-A5 acts as a repulsive cue for tangentially migrating neurons
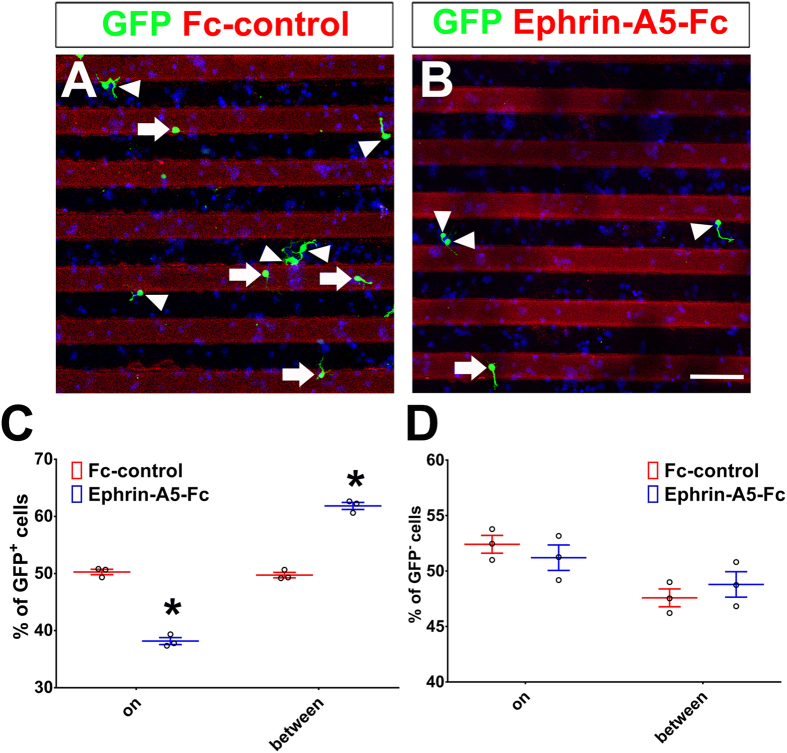
Since our results show that Ephb2 is a direct target of Ascl1 and is required for the separation of the VZ/SVZ and IZ routes, EphB2 interacting Ephrin(s) should be present in specific areas of the dorsal telencephalon to repel tangentially migrating cortical interneurons. Previous studies have shown that Ephrin-A5 and Ephrin-B2 potentially interact with EphB233,34. Ephrin-A5 (Efna5) is expressed in the MGE and preoptic area (POA) of the ventral telencephalon and acts as a repulsive cue for interneurons35. Efna5 mRNA and Ephrin-A5 protein have been detected in the CP of the dorsal telencephalon36,37. We examined the distribution of Ephrin-A5 in the telencephalon of E19.5 rat embryo by immunofluorescence. High levels of Ephrin-A5 were detected in deep VZ, upper SVZ, deep IZ and CP, but the signal was lower in the VZ/SVZ and IZ routes for tangential migration (Fig. S11). This result suggests that Ephrine-A5 may serve as a repulsive cue for cortical interneurons. To test this hypothesis, recombinant Ephrin-A5-Fc chimera (Ephrin-A5-Fc) or control protein (Fc-control) was coated onto 50 μm-wide stripes with Alexa 549 donkey-anti-guinea pig antibody for visualizing the coated area. Expression constructs of Ascl1 and GFP were co-electroporated into the dorsal telencephalon. Cells near the electroporated site were dissected two days after electroporation and cultured on the coverslips with coated stripes. 16 to 18 hours after plating, GFP-negative cells were equally distributed on stripes and between stripes of Fc-control or Ephrin-A5-Fc (Fig. 7D). While Ascl1-expressing GFP-positive cells were distributed equally on and between Fc-control stripes, they were preferentially distributed between Ephrin-A5-Fc stripes (Fig. 7A–C; mean ± SEM = 49.7 ± 0.8% in Fc-control group, 61.8 ± 1.1% in Ephrin-A5-Fc group, n = 3) and avoided the Ephrin-A5-Fc coated stripes (Fig. 7A–C; 50.3 ± 0.8% in Fc-control group, 38.2 ± 1.1% in Ephrin-A5-Fc group, n = 3). This result suggests that Ephrin-A5 acts as a repulsive cue for Ascl1-expressing cells.
Figure 7. Ephrin-A5 has a repulsive effect on Ascl1-expressing cortical neurons.
Control (US2) or Ascl1 expression constructs were electroporated into the dorsal telencephalon of E15.5 rats. The dorsal telencephalon was dissected two days after electroporation and dissociated into individual cells. These cells were cultured on coverslips coated with Fc-control or Ephrin-A5-Fc stripes. Cells were fixed 16–18 hours after plating and cells on strips or between strips were counted. Electroporated cells were labeled with anti-GFP in green, nuclear DNA was stained with DAPI in blue. GFP-positive cells on stripes are indicated by white arrowheads; GFP-positive cells between stripes are indicated by white arrows. (A) On a coverslip coated with Fc-control, GFP-positive Ascl1-expressing cells and GFP-negative cells were distributed evenly on stripes and between stripes. (B) On a coverslip coated with Ephrin-A5-Fc, GFP-positive Ascl1-expressing cells were preferentially distributed between stripes, while GFP-negative cells were evenly distributed on stripes and between stripes. Length of the scale bar is 50 μm. (C,D) Distribution of GFP-positive and GFP-negative cells. 150 cells from each group were counted in each experiment. Data are presented as mean ± SEM with all data points and analyzed by using Chi-square test, n = 3. *p < 0.05 compared to the Fc-control.
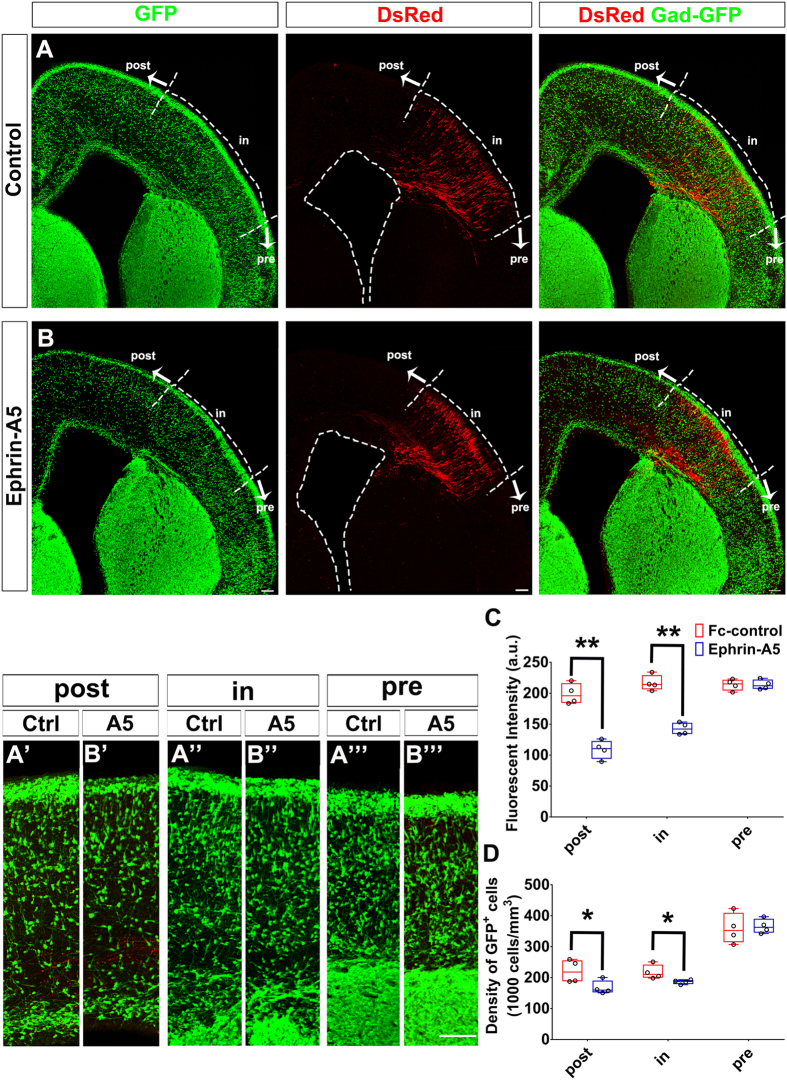
To investigate whether Ephrin-A5 repulses tangentially migrating cells in vivo, we examined interneuron distribution in Gad67-GFP mice. Expression constructs of Efna5 and DsRed were co-electroporated into the dorsal telencephalon at E14.5. Based on our hypothesis, these Efna5-expressing DsRed-positive cells should repel migrating GFP-positive cortical interneurons. In comparison with the control group, fewer GFP-positive cortical interneurons were observed dorsomedial to the region containing DsRed-positive Efna5-expressing cells (Fig. 8A,B). We quantified the average fluorescent intensity and the density of GFP-positive cells in the pre-, in-, or post-DsRed region (Fig. 8A,B). In comparison with the control group, the average fluorescent intensity of the mental zone (MZ) was reduced in the post-DsRed (Fig. 8C; control: 198.9 ± 8.4, Ephrin-A5: 109.1 ± 7.6; n = 4) and in-DsRed regions (Fig. 8C; control: 216.6 ± 6.3, Ephrin-A5: 142.8 ± 4.9; n = 4) when Efna5 was over-expressed. The density of GFP-positive cells of the CP, IZ, SVZ/VZ was also reduced in the post-DsRed region when Efna5 was over-expressed (Fig. 8C; post-DsRed, control: 2.2 × 105 ± 1.9 × 104 cells/mm3, Ephrin-A5: 1.7 × 105 ± 1.1 × 104 cells/mm3; n = 4). This result indicates that Ephrin-A5 repels tangentially migrating cortical interneurons and is likely to act in the cortex to separate the VZ/SVZ and IZ routes.
Figure 8. Ephrin-A5 repulses cortical interneurons.
Control (US2) or EfnA5 expression constructs were electroporated into the dorsal telencephalon together with a DsRed expression construct at E14.5 of Gad67-GFP mice. Brains were dissected four days after electroporation and sectioned coronally. (A,B) Confocal image of E18.5 mouse telencephalon. The cortex was divided into three regions: (A”,B”) the “in” region that contained DsRed-positive cells; (A”’,B”’) the “pre” region was ventrolateral to the “in” region; (A’,B’) the “post” region was dorsomedial to the “in” region. (A,B) In Ephrin-A5 group, fewer GFP-positive cells were present in “post” regions than those of the control group. Length of the scale bar is 100 μm. (C) Quantification of GFP fluorescent intensity in the MZ. (D) Quantification of GFP-positive cells in the CP, IZ and VZ/SVZ. A 200 μm wide cortical area in each region was selected for quantification. For the pre-DsRed, we selected the 200 μm wide cortical area 50 to 100 μm from the pre-/in-DsRed boundary. For the in-DsRed, we selected 50 to 100 μm from the in-/post-DsRed boundary. For the post-DsRed, we selected 50 to 100 μm from the in-/post-DsRed boundary. Data are presented as box and whisker plots and analyzed by using Student’s t-test, n = 4. *p < 0.05.
Discussion
Although Ascl1 is required for the generation of some cortical interneurons, whether Ascl1 promotes tangential migration has never been revealed. Here, we demonstrate that both Ascl1 and Dlx2 promote tangential migration of neurons derived from either ventral or dorsal telencephalon (Figs 1 and 2). Ascl1 promotes tangential migratory routes through the VZ/SVZ and IZ of the dorsal telencephalon in a Dlx2-dependent and a Dlx2-independent manner, respectively (Fig. 2). Furthermore, Ascl1 induces the expression of Ephb2 to confine migratory routes (Figs 4, 5, 6). Ephrin-A5 repels Ascl1-expressing neurons in vitro and cortical interneurons in vivo (Figs 7 and 8), suggesting that the separation of the two migratory routes is maintained by EphB2-Ephrin-A5 signaling. Thus, we propose that Ascl1 induces Ephb2 in interneurons to avoid Efna5-expressing areas and migrate along the VZ/SVZ and IZ routes (Fig. S12).
Several transcription factors regulate cell fate specification and neuronal migration in parallel. However, it is difficult to distinguish whether these transcription factors specify cell fate first and in turn regulate migration versus they play more direct roles in migration. Therefore, it is important to identify target genes of these transcription factors to confirm their roles in migration. Ascl1, Dlx1/2 and Nkx2.1 are important for cell fate specification in the ventral telencephalon. Loss of Ascl1 , Dlx1/2 or Nkx2 .1 attenuates tangential migration of interneurons13,19,23. Dlx1/2 repress the expression of Pak3 and Map2, two genes encoding regulators of cytoskeletal dynamics. Reduction of Pax3 and MAP2 in Dlx1/2 knockout mice rescues the tangential migration defect15, suggesting that Pax3 and MAP2 act downstream of Dlx2 in regulating migration. Dlx1/2 also repress the expression of Neuropilin 2 (Nrp2), which encodes a receptor for Semaphorin (Sema)-3A and 3F, to facilitate migration of interneurons toward the dorsal telencephalon21. Nkx2.1 induces ErbB4 in MGE-derived interneurons to enable them to be attracted to the striatum where Neuregulin 1 is expressed38. Nkx2.1 also induces Ephb1 and Ephb3 in striatal interneurons to prevent them from entering the dorsal telencephalon38. Here, we show that Ascl1 contributes to confine routes of tangential migration in the dorsal telencephalon through inducing EphB2. Taken together, these studies all support the model that transcription factors known to specify cell fates may also play a direct role in regulating cell migration.
Previous studies have demonstrated that cortical interneurons generated at different stages select different migratory routes in the dorsal telencephalon7. From E12 to E15, cortical interneurons migrate through the MZ and IZ of the dorsal telencephalon39,40,41. After E15, cortical interneurons migrate through the VZ/SVZ, IZ, and MZ. Interestingly, Ascl1 and Dlx1/2 control the generation of interneurons sequentially in the developing telencephalon13. In Ascl1 knockout mice, early-born interneurons at E10.5 and migrating interneurons in the dorsal telencephalon at E12.5 are decreased, suggesting that Ascl1 regulates generation of interneurons at early developmental stage from E10.5 to E12.519,42. On the other hand, interneurons born after E15.5 are decreased in Dlx1/2 knockout mice, suggesting that Dlx1/2 regulate interneuron generation at late developmental stages13,43. These data suggest that Ascl1 and Dlx2 promote different migratory routes in the dorsal telencephalon, which coincides with our findings. Ascl1 promotes tangential migration through the IZ, which is the major migratory route for early-born interneurons (Figs 1F and 2D). Either Ascl1 or Dlx2 promotes tangential migration through the VZ/SVZ route (Figs 1F,G and 2D,E), which is the main migratory route for late-born interneurons. While cortical interneurons are shown to tangentially migrate through the VZ/SVZ, IZ, and MZ of the dorsal telencephalon, the molecular mechanism underlying route selection has never been identified. Thus, our data is the first to provide evidence that Ascl1 and Dlx2 may be key for different migratory behaviors of early- versus late-born cortical interneurons.
Ascl1 has been suggested to act upstream of Dlx1/2 during development of the ventral telencephalon20. Interestingly, Ascl1 may regulate the expression of Dlx1/2 negatively or positively44,45. In the ventral telencephalon, Ascl1 maintains the pool of neural progenitors at least in part by regulating the Notch pathway42. Loss of Ascl1 leads to premature neuronal differentiation and expansion of Dlx1/2 expression domain19,42. In the developing thalamus, Ascl1 represses Dlx1/2 expression through Helt, a bHLH-Orange factor, to specify GABAergic neurons from thalamic progenitors45. These findings suggest that Ascl1 acts as a negative regulator for Dlx1/2 expression in some cellular contexts. Here, we found that expression of Dlx2 was increased by expression of Ascl1 in P19 cells (Fig. S6A), consistent with previous findings that Ascl1 activates Dlx1/2 in the ventral telencephalon17,20. Ascl1 and Dlx1/2 have also been demonstrated to act in parallel to regulate neuronal differentiation and migration of LGE-derived olfactory interneurons46. Since knockdown of Dlx2 attenuates the VZ/SVZ migratory route promoted by Ascl1 (Fig. 2F,G), and simultaneous expression of Ascl1 and Dlx2 promotes migration through the VZ/SVZ (Fig. S13), our results suggest that Ascl1 induces Dlx2 to promote tangential migration of interneurons through the VZ/SVZ route of the dorsal telencephalon. In addition, while expression of Ascl1 induced Ephb2 in P19 cells, expression of Dlx2 also increased the level of Ephb2 mRNA (3.0 ± 0.9 fold of the control, n = 4, Fig. 4). Comparing with expression of Dlx2 alone, simultaneous expression of Dlx2 and knockdown of Ephb2 decreased GFP-positive cells in the VZ/SVZ and increased cells in the IZ and CP (Fig. S14), demonstrating that EphB2 is also critical to confine the VZ/SVZ migratory route promoted by Dlx2. Together, our data suggest that Ascl1 acts upstream or cooperatively with Dlx2 to maintain the tangentially migratory route in the VZ/SVZ, possibly through inducing Ephb2 cooperatively.
For guiding tangential migration, a chemokine Cxcl12 is expressed in the dorsal telencephalon to maintain interneurons in a specific migratory route. Cxcl12 is expressed in the VZ/SVZ to attract cortical interneurons expressing Cxcr4 and Cxcr7 receptor genes47,48,49,50. Here, we report that migrating interneurons are confined to distinct migratory routes by EphB2/Ephrin-A5 signaling. Thus, chemo-attractive and -repulsive cues may coordinate to guide tangential migration of cortical interneurons.
Here we identify Ephb2 as a direct target of Ascl1. This is supported by a previous study using ChIP-sequencing in a cellular model of neurogenesis driven by over-expressed Ascl151. Previous studies have also reported that Ascl1 regulates radial migration in glutamatergic neurons by inducing a small GTPase gene Rnd3 and Cenpj, a gene encoding a centrosome interacting protein, in the dorsal telencephalon11,16. Here, we show that Ascl1 is sufficient to promote tangential migration even in the dorsal telencephalon. How does Ascl1 affect different migratory programs in different types of neurons? One possibility is that high level of Ascl1 is capable to activate the tangentially migratory program since Ascl1 is expressed strongly in the ventral telencephalon but only at a relatively low level in the dorsal telencephalon52. Another possibility is that Ascl1 coordinates with other transcription factors or signaling pathways in different cells to determine the migratory program.
In this study, we provide evidence that Ascl1 promotes tangential migration and begin to unravel the mechanisms that underlie this phenomenon. It will be interesting to identify additional Ascl1 target genes that contribute to the regulation of tangential migration. In addition to the links between interneurons and some neurological and psychological diseases1,2, interneuron transplantation has been demonstrated to relieve seizures, symptoms of Parkinson’s disease and neuropathic pain53. A better understanding of the regulation of interneuron migration may contribute to therapeutic approaches for diseases related to interneuron dysfunction.
Methods
Animals
Timed-pregnant Sprague Dawley (SD) rats were obtained from the Laboratory Animal Center in the National Yang-Ming University (NYMU). The Gad67-GFP-knock-in (Gad67-GFP) mice were provided by Dr. Yanagawa24,54. All animals were housed in the LAC in NYMU and handled according to the guidelines and animal use protocols approved by the Institutional Animal Care and Use Committee (Approval #: 1031246). Gad67-GFP mice were maintained as heterozygous. The sequences of PCR primers used for Genotyping of Gad67-GFP mice are: 5′-GGCACAGCTCTCCCTTCTGTTTGC, GCTCTCCTTTCGCGTTCCGACAG, and CTGCTTGTCGGCCATGATATAGACG. The PCR product was a 564-base pair (bp) fragment for the Gad67-GFP allele and a 265-bp fragment for the wildtype allele. The embryonic stage was determined as E0.5 for the day when the vaginal plug was observed.
Plasmids
sh-LacZ#1 , sh-LacZ#2 , sh-Dlx2#1 , shDlx2#2 , shEphb1#1 , shEphb1#2 , shEphb2#1, and shEphb2#2 are in pLKO shRNA expression vector from The RNAi Consortium (TRC) shRNA Library at the Broad Institute. Target sequences of these shRNA constructs are listed in Table S1. Mouse Neurog2 (NM_009718), Ascl1 (NM_008553), Dlx2 (NM_010054), and Efna5 (NM_207654) were amplified by PCR and inserted into the US2 vector with human Ubiquitin C promoter55,56.
In utero electroporation
Surgery for in utero electroporation was performed in E15.5 rats and E14.5 mice. The procedure has been described previously57. Animals were anesthetized with 2.5% isoflurane (Sigma-Aldrich). An incision was made through the skin and abdominal muscle to expose the viscera. The uterine horns were carefully exposed and placed on wet gauze. DNA solution (2 μg/μL, 0.5 μl total volume) was injected into left lateral ventricle of the telencephalon for each embryo. Forceps electrodes (7 mm in diameter for rats and 5 mm for mice; Harvard Apparatus) were used and five electric pulses separated by 500 ms were transmitted at 50 V for rats and 40 V for mice by an electroporation generator (Harvard Apparatus). The uterine horns were put back into the abdominal cavity after electroporation. Embryos were harvested four days after electroporation for experiments in Figs 1,2,5 and 8, S1, S2, S3, S4, S5, S7, S8, S12 and S13; two days for experiments in Figs 3 and 6, S4 and Supplementary videos.
Live imaging
Coronal slices of embryonic rat brains were prepared 48 hours after electroporation. Slices were placed on Millicell-CM inserts (Millipore) and incubated at 37 °C with 5% CO2 in culture medium that containing 25% Hanks balanced salt solution, 47% basal MEM (Invitrogen), 25% normal horse serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen), and 0.66% glucose. Multiple GFP-positive cells were imaged on an inverted microscope. Time-lapse images were captured by using camera (CoolSNAP HQ; Roper Scientific) at intervals of six minutes for 5 hours and data were analyzed by using MetaMorph software (Molecular Devices).
Fixation and sectioning
Embryos were perfused with saline and then 4% paraformaldehyde (PFA, Sigma). Brains were post-fixed in 4% PFA overnight, then cryoprotected with 30% sucrose solution and cut frozenly into 60 μm coronal sections with a sliding microtome (Leica). Brain sections were collected and storage in the PBS with 0.02% sodium azide (Sigma) at 4 °C.
Transfection and differentiation of P19 cells
Mouse P19 cells were maintained in αMEM (Gibco) medium supplement with 100 units/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), 7.5% fetal bovine serum (Hyclone), and 2.5% calf serum (Hyclone). For transfection, cells were plated in 6-well dishes at 80–90% confluence without antibiotics. Lipofectamine 2000 (Invitrogen) was used for transfection according to the manufacturer’s instruction.
Stripe assay and primary culture of cortical neurons
The procedure of stripe assay has been described previously58. A silicon wafer was used to generate a template for poly(dimethylsiloxane) (PDMS) molds with parallel 50 μm stripes separated by 50 μm gaps. PDMS mold was reversibly sealed on 22 × 22 mm2 coverslip to form channels for microfluid injection. Microfluid composed of Alexa 549-conjugated donkey anti-guinea pig IgG (1:50, Abcam) and Fc-control (3 μg/ml, Enzo) or Ephrin-A5-Fc (4 μg/ml, R&D system). After injection, microfliud passed through channels by suction. Coverslips were dried for 16 to 18 hours at room temperature and the alternative stripes were formed. Poly-L-lysine (Sigma-Aldrich) and laminin (Sigma-Aldrich) were coated 16 to 18 hours before use.
The protocol for culturing cortical neurons has been described previously28. Two days after in utero electroporation, the dorsal telencephalon was dissected form E17.5 rat embryos and dissociated by trituration into single cell suspension with fire-polished Pasteur pipettes. 5 × 106 cells were cultured on a coverslip with stripe pattern in L15 medium (Gibco) with N2, B27 (Invitrogen), 30 mM Glucose, 26 mM NaHCO3, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Cells were cultured for 16 to 18 hours and fixed in 4% PFA. At least 150 GFP-positive neurons were counted for each group.
Immunofluorescence
Sections or coverslips with cells were washed with Tris buffered saline (TBS) and incubated in blocking buffer (1% glycine, 0.4% Triton X-100, 3% BSA, 0.1% sodium azide and 10% normal goat serum in TBS) for one hour at room temperature. Sections or coverslips were incubated in the primary antibodies in species-appropriate combinations for 24 hours at 4 °C. Primary antibodies and dilution used in our experiments included rabbit anti-GFP (Catalog: A-11122, 1:1500, Invitrogen), rat anti-GFP (Catalog: GF090R, 1:2000, Nacalai), mouse anti-MAP2 (Catalog: ab11267, 1:200, Abcam), Goat anti-EphB2 (Catalog: AF467, 1:250, R&D systems), rabbit anti-Ephrin-A5 (Catalog: ab70114, 1:250, abcam) and rabbit anti-RFP (Catalog: ab62341, 1:250, Abcam). After incubation with primary antibodies, sections or coverslips were washed with TBS and incubated with secondary antibodies in blocking buffer at room temperature for two hours. Secondary antibodies included Alexa 488, 546, 633 conjugated goat anti-mouse, anti-rat, anti-rabbit IgG (1:500; Abcam). Sections or coverslips were then washed with TBS and mounted with ProLong Gold anti-fade (Invitrogen). All images were taken by Zeiss LSM700 confocal microscopy. The volume, migratory distance, and the average fluorescent intensity were estimated by using ImageJ (U. S. National Institutes of Health).
RNA extraction and Quantitative RT-PCR
P19 cells in 6-well plates were transfected with 1 μg of US2-puro (puromycin-resistant gene) and 3 μg of various expression constructs. Transfected cells were selected in puromycin (15 μg/ml) for eight hours. Total RNA was extracted one day after transfection by using the RNeasy Mini Kit (QIAGEN). cDNA was prepared from 3 μg of total RNA by using SuperScript III Reverse Transcriptase (Invitrogen). 10 μl of PCR mix contains 1× FastStart Universal SYBR Green Master (Roche), 20 μM forward primer, 20 μM reverse primer (primer sequences are listed in Table S2), and 2.5 ng/μl cDNA template. Quantitative PCR was performed with StepOnePlus™ Real-Time PCR System (Applied Biosystems) and analyzed with the StepOnePlus V2.3 software. TATA-box binding protein (Tbp) was used as a control. MIQE is in Table S3 and raw data of qPCR are available in Supplementary spreadsheet I.
Chromatin Immunoprecipitation (ChIP)
The procedure of ChIP has been described previously59. P19 cells were transfected with Ascl1 expression vectors and cultured for one day. Genomic DNA from 1 × 107 cells was used for precipitation with each antibody. Cellular contents were cross-linked with 1% formaldehyde for five minutes at room temperature and stopped by incubation in 0.125 M of glycine for 10 minutes at room temperature. Fixed cells were rinsed twice with PBS and re-suspended by using Trypsin-EDTA (Invitrogen). Cell lysate was then sonicated for 5 min and centrifuged at 14000 rpm for 10 min. The cleared supernatant was used immediately for IP. Sonicated DNA was blocking for one hour at room temperature and incubated with mouse anti-Ascl1 (BD Biosciences) or rabbit non-immune serum IgG (Alpha Diagnostic International) for overnight. Precipitated materials were eluted by elution buffer (SDS 1%, 0.1 M NaHCO3). DNA was reverse-cross-linked in 5M NaCl and treated with RNase and proteinase K. DNA was purified by using phenol-chloroform. PCR primer sequences for amplifying putative Ascl1 binding fragments were listed in the Table S3. Raw data of qPCR are available as Supplementary spreadsheet II.
Statistical analysis
Statistical analysis was performed by using SPSS (IBM). Two-tailed Student’s t test was used for comparison between two groups and ANOVA with Tukey’s-HSD post hoc test was used for comparison among three or more groups. Chi-square analysis was used for the analysis in Fig. 6.
Additional Information
How to cite this article: Liu, Y.-H. et al. Ascl1 promotes tangential migration and confines migratory routes by induction of Ephb2 in the telencephalon. Sci. Rep. 7, 42895; doi: 10.1038/srep42895 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr. Cheng-Chang Lien for helping Gad67-GFP mouse breeding, Dr. Ming-Ji Fann and Dr. Shen-Ju Chou for discussion of the manuscript. We thank Imaging Core of NYMU for imaging analysis. RNAi reagents were obtained from the National Core Facility for Manipulation of Gene Function by RNAi, miRNA, miRNA sponges, and CRISPR/Genomic Research Center, Academia Sinica, supported by the National Core Facility Program for Biotechnology Grants of Ministry of Science and Technology (MOST), Taiwan, 204-2319-B-001-001. This research is funded by MOST Grants 101-2311-B-010-007-MY3, 104-2321-B-010-018, 104-2311-B-010-004, 104-2320-B-003-008 and Brain Research Center, NYMU.
Footnotes
The authors declare no competing financial interests.
Author Contributions Yuan-Hsuan Liu designed and performed the experiments, analyzed the data, and wrote the manuscript. Jin-Wu Tsai designed and performed the experiments, and analyzed the data. Jia-Long Chen performed the experiments. Wan-Shan Yang performed the experiments and analyzed the data. Pei-Ching Chang designed the experiments and analyzed the data. Pei-Lin Cheng designed and performed the experiments, and analyzed the data. David L. Turner designed the experiments and analyzed the data. Yuchio Yanagawa designed the experiments. Tsu-Wei Wang designed the experiments, analyzed the data, and wrote the manuscript. Jenn-Yah Yu designed and performed the experiments, analyzed the data, and wrote the manuscript.
References
- Rossignol E. Genetics and function of neocortical GABAergic interneurons in neurodevelopmental disorders. Neural Plast 2011, 649325, doi: 10.1155/2011/649325 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci 13, 107–120, doi: 10.1038/nrn3155 (2012). [DOI] [PubMed] [Google Scholar]
- Marin O. & Rubenstein J. L. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci 2, 780–790, doi: 10.1038/35097509 (2001). [DOI] [PubMed] [Google Scholar]
- Xu Q., Cobos I., De La Cruz E., Rubenstein J. L. & Anderson S. A. Origins of cortical interneuron subtypes. J Neurosci 24, 2612–2622, doi: 10.1523/JNEUROSCI.5667-03.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt S. J. et al. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron 48, 591–604, doi: 10.1016/j.neuron.2005.09.034 (2005). [DOI] [PubMed] [Google Scholar]
- Kwan K. Y., Sestan N. & Anton E. S. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development 139, 1535–1546, doi: 10.1242/dev.069963 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka D. H. & Nakajima K. Migratory pathways of GABAergic interneurons when they enter the neocortex. Eur J Neurosci 35, 1655–1660, doi: 10.1111/j.1460-9568.2012.08111.x (2012). [DOI] [PubMed] [Google Scholar]
- Guo J. & Anton E. S. Decision making during interneuron migration in the developing cerebral cortex. Trends Cell Biol 24, 342–351, doi: 10.1016/j.tcb.2013.12.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M., Stoykova A. & Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21, 1031–1044 (1998). [DOI] [PubMed] [Google Scholar]
- Parras C. M. et al. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev 16, 324–338, doi: 10.1101/gad.940902 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacary E. et al. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron 69, 1069–1084, doi: 10.1016/j.neuron.2011.02.018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W. et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci USA 103, 1319–1324, doi: 10.1073/pnas.0510419103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O., Anderson S. A. & Rubenstein J. L. Origin and molecular specification of striatal interneurons. J Neurosci 20, 6063–6076 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega-Pereira S. et al. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron 59, 733–745, doi: 10.1016/j.neuron.2008.07.024 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I., Borello U. & Rubenstein J. L. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron 54, 873–888, doi: 10.1016/j.neuron.2007.05.024 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez P. P. et al. Cenpj/CPAP regulates progenitor divisions and neuronal migration in the cerebral cortex downstream of Ascl1. Nat Commun 6, 6474, doi: 10.1038/ncomms7474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C. et al. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev 14, 67–80 (2000). [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G. et al. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci 30, 1582–1594, doi: 10.1523/JNEUROSCI.4515-09.2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S., Fode C. & Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development 126, 525–534 (1999). [DOI] [PubMed] [Google Scholar]
- Poitras L., Ghanem N., Hatch G. & Ekker M. The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development 134, 1755–1765, doi: 10.1242/dev.02845 (2007). [DOI] [PubMed] [Google Scholar]
- Le T. N. et al. Dlx homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J Biol Chem 282, 19071–19081, doi: 10.1074/jbc.M607486200 (2007). [DOI] [PubMed] [Google Scholar]
- Colasante G. et al. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J Neurosci 28, 10674–10686, doi: 10.1523/JNEUROSCI.1283-08.2008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. A., Eisenstat D. D., Shi L. & Rubenstein J. L. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474–476 (1997). [DOI] [PubMed] [Google Scholar]
- Tamamaki N. et al. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol 467, 60–79, doi: 10.1002/cne.10905 (2003). [DOI] [PubMed] [Google Scholar]
- Rodger J., Salvatore L. & Migani P. Should I stay or should I go? Ephs and ephrins in neuronal migration. Neurosignals 20, 190–201, doi: 10.1159/000333784 (2012). [DOI] [PubMed] [Google Scholar]
- North H. A., Clifford M. A. & Donoghue M. J. ‘Til Eph do us part’: intercellular signaling via Eph receptors and ephrin ligands guides cerebral cortical development from birth through maturation. Cereb Cortex 23, 1765–1773, doi: 10.1093/cercor/bhs183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah M. H. et al. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127, 693–702 (2000). [DOI] [PubMed] [Google Scholar]
- Yu J. Y., Wang T. W., Vojtek A. B., Parent J. M. & Turner D. L. Use of short hairpin RNA expression vectors to study mammalian neural development. Methods Enzymol 392, 186–199, doi: 10.1016/S0076-6879(04)92011-3 (2005). [DOI] [PubMed] [Google Scholar]
- Zimmer G. et al. Bidirectional ephrinB3/EphA4 signaling mediates the segregation of medial ganglionic eminence- and preoptic area-derived interneurons in the deep and superficial migratory stream. J Neurosci 31, 18364–18380, doi: 10.1523/JNEUROSCI.4690-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M. E. & Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20, 429–440 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro D. S. et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev 25, 930–945, doi: 10.1101/gad.627811 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke R. M., Meredith D. M., Borromeo M. D., Savage T. K. & Johnson J. E. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev Biol 328, 529–540, doi: 10.1016/j.ydbio.2009.01.007 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen J. P. et al. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci 7, 501–509, doi: 10.1038/nn1237 (2004). [DOI] [PubMed] [Google Scholar]
- Lin K. T., Sloniowski S., Ethell D. W. & Ethell I. M. Ephrin-B2-induced cleavage of EphB2 receptor is mediated by matrix metalloproteinases to trigger cell repulsion. J Biol Chem 283, 28969–28979, doi: 10.1074/jbc.M804401200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer G. et al. Ephrin-A5 acts as a repulsive cue for migrating cortical interneurons. Eur J Neurosci 28, 62–73, doi: 10.1111/j.1460-9568.2008.06320.x (2008). [DOI] [PubMed] [Google Scholar]
- Mackarehtschian K., Lau C. K., Caras I. & McConnell S. K. Regional differences in the developing cerebral cortex revealed by ephrin-A5 expression. Cereb Cortex 9, 601–610 (1999). [DOI] [PubMed] [Google Scholar]
- Deschamps C. et al. EphrinA5 protein distribution in the developing mouse brain. BMC Neurosci 11, 105, doi: 10.1186/1471-2202-11-105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Cervino V. et al. Molecular mechanisms controlling the migration of striatal interneurons. J Neurosci 35, 8718–8729, doi: 10.1523/JNEUROSCI.4317-14.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N., Fujimori K. E. & Takauji R. Origin and route of tangentially migrating neurons in the developing neocortical intermediate zone. J Neurosci 17, 8313–8323 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. A., Marin O., Horn C., Jennings K. & Rubenstein J. L. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development 128, 353–363 (2001). [DOI] [PubMed] [Google Scholar]
- Antypa M., Faux C., Eichele G., Parnavelas J. G. & Andrews W. D. Differential gene expression in migratory streams of cortical interneurons. Eur J Neurosci 34, 1584–1594, doi: 10.1111/j.1460-9568.2011.07896.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K. et al. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development 129, 5029–5040 (2002). [DOI] [PubMed] [Google Scholar]
- Anderson S. A. et al. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19, 27–37 (1997). [DOI] [PubMed] [Google Scholar]
- Petryniak M. A., Potter G. B., Rowitch D. H. & Rubenstein J. L. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron 55, 417–433, doi: 10.1016/j.neuron.2007.06.036 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. et al. Ascl1 and Helt act combinatorially to specify thalamic neuronal identity by repressing Dlxs activation. Dev Biol 398, 280–291, doi: 10.1016/j.ydbio.2014.12.003 (2015). [DOI] [PubMed] [Google Scholar]
- Long J. E. et al. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci 27, 3230–3243, doi: 10.1523/JNEUROSCI.5265-06.2007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm R. K. et al. CXCR4 regulates interneuron migration in the developing neocortex. J Neurosci 23, 5123–5130 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubedo N., Cerdan E., Sapede D. & Rossel M. CXCR4 and CXCR7 cooperate during tangential migration of facial motoneurons. Mol Cell Neurosci 40, 474–484 (2009). [DOI] [PubMed] [Google Scholar]
- Sanchez-Alcaniz J. A. et al. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron 69, 77–90, doi: 10.1016/j.neuron.2010.12.006 (2011). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron 69, 61–76, doi: 10.1016/j.neuron.2010.12.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo A. A. et al. Ascl1 Coordinately Regulates Gene Expression and the Chromatin Landscape during Neurogenesis. Cell Rep, doi: 10.1016/j.celrep.2015.02.025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britz O. et al. A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex 16 Suppl 1, i138–151, doi: 10.1093/cercor/bhj168 (2006). [DOI] [PubMed] [Google Scholar]
- Southwell D. G. et al. Interneurons from embryonic development to cell-based therapy. Science 344, 1240622, doi: 10.1126/science.1240622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang P. H., Chien T. C., Chen C. C., Yanagawa Y. & Lien C. C. ASIC-dependent LTP at multiple glutamatergic synapses in amygdala network is required for fear memory. Sci Rep 5, 10143, doi: 10.1038/srep10143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. H. et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res 34, e53, doi: 10.1093/nar/gkl143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. Y., Chung K. H., Deo M., Thompson R. C. & Turner D. L. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res 314, 2618–2633, doi: 10.1016/j.yexcr.2008.06.002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. W., Chen Y., Kriegstein A. R. & Vallee R. B. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol 170, 935–945, doi: 10.1083/jcb.200505166 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P. L., Lu H., Shelly M., Gao H. & Poo M. M. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron 69, 231–243, doi: 10.1016/j.neuron.2010.12.021 (2011). [DOI] [PubMed] [Google Scholar]
- Yang W. S., Hsu H. W., Campbell M., Cheng C. Y. & Chang P. C. K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation. PLoS Pathog 11, e1005051, doi: 10.1371/journal.ppat.1005051 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.