Figure 3. Two TFAM can bind a single tetramolecular G4, which is not recognized like B-DNA.
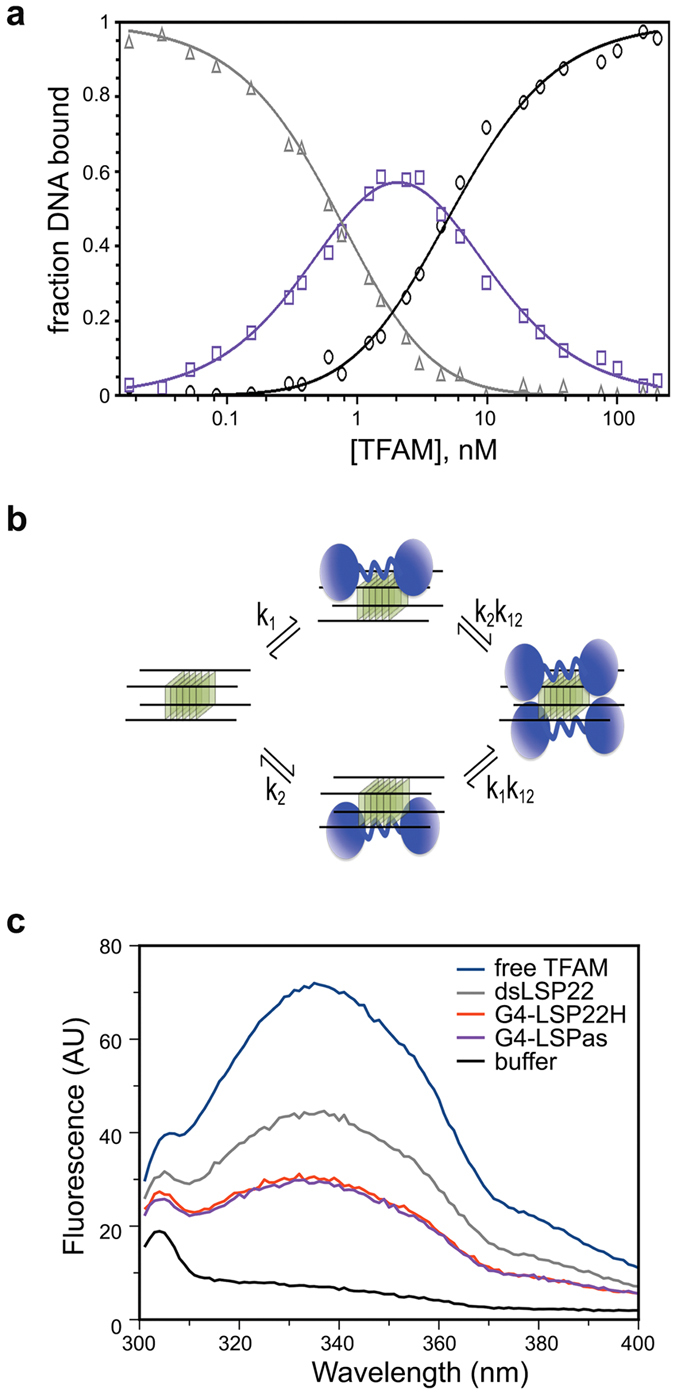
(a) Gel-mobility shift titration for all types of complexes upon TFAM binding to G4-LSP22H (Fig. 2e). Fractions of unbound (triangles), singly bound (shift I, squares), and doubly bound (shift II, circles) DNA are represented. The solid curves result from fitting the data according to Eqs 3 (b) Model of binding of two TFAM molecules (in blue) to two sites on a tetramolecular G4. The intrinsic association constants, k1 and k2, represent the binding constants to G4 sites 1 and 2, while k12 is the cooperativity parameter representing the increased stability of protein-DNA complexes resulting from binding two protein molecules to the two G4 sites. (c) Intrinsic fluorescence emission spectra of TFAM (0.5 μM) in the absence (blue) and presence of 1.5 μM dsLSP22 (grey), G4-LSPas (violet) and G4-LSP22H (red) at an excitation wavelength of 275 nm.
