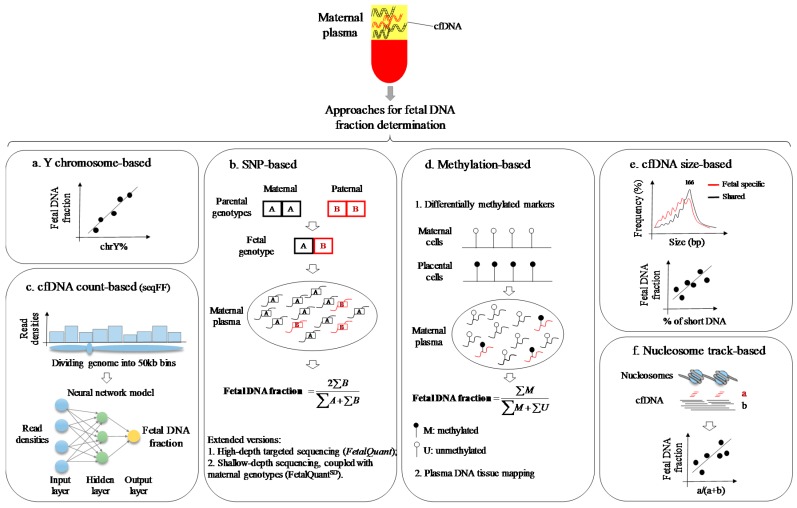
Figure 1.
Schematic illustration of current approaches for the determination of fetal DNA fraction in maternal circulating cell-free DNA (cfDNA). (a) Y chromosomal (chr) sequence-based fetal DNA fraction estimate [3,22]; (b) Single-nucleotide polymorphism (SNP)-based approach. A direct way to estimate the fetal DNA fraction is to use the SNP loci, where both mother and father are homozygous but with different alleles. The resulting fetal genotype is obligately heterozygous. In maternal plasma, the fetal DNA fraction can be directly deduced by calculating the proportion of fetal specific alleles [9,30]. Based on this concept, two extended versions of SNP-based methods for fetal DNA fraction estimate have been developed, namely FetalQuant and FetalQuantSD, which can be used without the need of both paternal and maternal genotype information [31,32]; (c) cfDNA count-based approach. Read densities across the genome-wide 50 KB windows are fitted into a neural network model to predict the fetal DNA fraction [33]; (d) Differential methylation-based approaches [17,26,34,35]; (e) cfDNA size-based approach. The proportion of short cfDNA molecules is correlated with fetal DNA fraction [36]; (f) Nucleosome track-based approach. Cell-free DNA distribution at the nucleosomal core and linker regions is correlated with fetal DNA fraction [37].