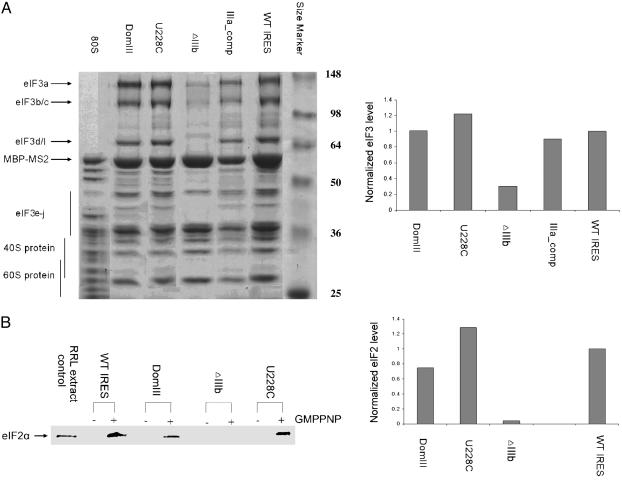
Fig. 3.
Analysis of initiation factor presence within translation complexes assembled on wild-type and mutant HCV IRES RNAs. (A Left) Coomassie blue-stained 10% SDS/PAGE gel of affinity-purified 48S and 80S samples; IRES constructs are indicated at the top, protein size markers are labeled at right, and bands corresponding to eIF3, 40S, and 60S proteins are indicated at left. The identities of eIF3 bands were verified by Western blotting (data not shown). (Right) Relative eIF3 levels in wild-type IRES and mutant IRES-bound complexes. For each sample, three eIF3 bands were quantitated by using imagej software, and the summed intensities were divided by the intensity of the MBP-MS2 band as a loading control; all values were normalized to that of the wild-type IRES-containing sample. Each value shown is the average of two independent experiments. (B Left) Western blot analysis of 48S complexes in the absence and presence of the nonhydrolyzable GTP analog GMPPNP using an anti-eIF2α antibody. Equal amounts of sample based on OD260 were applied to each lane. (Right) Relative eIF2 levels in wild-type and mutant IRES-bound complexes. Band intensities were quantitated by using imagej software and divided by the intensity of ribosomal protein bands in each lane to control for sample loading differences; all values were normalized to that of the wild-type IRES-containing sample. Each value shown, except for U228C, is the average of two independent experiments.