Abstract
Purpose
Emerging evidence has demonstrated that gut microbiome plays essential roles in the pathogenesis of human diseases in distant organs. Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by progressive loss of motor neurons. Treatment with the only FDA approved drug, Riluzole, extends patient life span only for a few months. Thus, there is an urgent need to develop novel interventions for alleviating the disease progression and improving the quality of life for ALS patients. Here we present evidence that intestinal dysfunction and dysbiosis may actively contribute to ALS pathophysiology.
Methods
We used G93A transgenic mice as a model of human ALS. The G93A mice show abnormal intestinal microbiome and damaged tight junctions, which occur before ALS disease onset. Feeding of the G93A mice with butyrate, a natural bacterial product.
Results
Feeding of the G93A mice with butyrate, restores the intestinal microbial homeostasis, improves the gut integrity, and prolongs the life span of the ALS mice. Paneth cells are specialized intestinal epithelial cells that regulate the host-bacterial interactions. At the cellular level, we show that abnormal Paneth cells were significantly decreased in the ALS mice treated by butyrate. Butyrate treatment decreases aggregation of the G93A-SOD1 mutated protein in both ALS mice and cultured human intestinal epithelial cells.
Implications
Our study highlights the complex role of gut microbiome and intestinal epithelium in the progression of ALS and presents butyrate as a potential therapeutic reagent to restore ALS dysbiosis.
Keywords: ALS, butyrate, dysbiosis, intestinal permeability, microbiome, tight junctions
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease. Most ALS patients die within three to five years due to respiratory paralysis.1 The lifetime risk of ALS is about 1 in 472 in women and 1 in 350 in men. Since ALS is an age-dependent disease, as the U.S. population increases and ages, an increase in the prevalence of ALS can be anticipated. Currently there is no intervention that can significantly change the course of the disease.2 Treatment with the only FDA approved drug, Riluzole, extends patient life span only for a few months. Thus, there is an urgent need to develop novel interventions for alleviating the disease progression and improving the quality of life for ALS patients.
Many cases of familial ALS (20–25%) are associated with mutations in the Cu/Zn superoxide dismutase gene (SOD1). An ALS mouse model G93A mice harbor human ALS-causing SOD1 mutations that recapitulate the neuronal and muscle impairment of human ALS patients. These mice are extensively used to investigate the pathomechanisms of ALS and trial new therapeutics.3–5 Our recent study revealed an exciting phenotype that linked aberrant microbial and intestinal homeostasis to disease progression in ALS mice.6 G93A mice have dysbiosis (imbalanced gut bacterial profile), disrupted tight junction structure, and increased intestinal permeability (leaky gut). These changes occur in young G93A mice before ALS symptom onset, suggesting that impaired gut-neuromuscular crosstalk may actively contribute to ALS progression and pathogenesis.
Intestinal microbiome community modulates numerous aspects of human physiology and is a critical factor in the development of chronic diseases.6 Intestinal epithelial cells are consistently exposed to bacteria, a process which plays a key role in development, renewal, and immunity.7–9 Frequent microbial challenges to epithelial cells trigger discrete signaling pathways, promoting molecular changes, such as the secretion of cytokines and chemokines, and alterations to molecules displayed at the epithelial cell surface. Intestinal homeostasis and microbiome play essential roles in neurological diseases, such as autism and Parkinson’s disease.3, 4 However, restoring intestinal homeostasis and gut microbiome in ALS is unexplored.
In the current study, we hypothesize that restoring microbiome and intestinal homeostasis delays disease onset and progression of ALS. A leaky gut can contribute to the altered microbiome environment that leads to reduced production of beneficial bacterial products. Indeed, our study has demonstrated that G93A mice show abnormal microbiome profile with reduced population of butyrate-producing bacteria. Remarkably, after being fed with the natural bacteria product, butyrate, the G93A mice exhibited a delay in onset of ALS symptoms, and a prolonged life span. Our current study has discovered butyrate for restoring intestinal homeostasis and microbiome, thus opening a new avenue in targeting the gut microbiota-butyrate-axis for ALS treatment.
Materials and Methods
Animals
G93A5 and age matched wild type mice were used in this study. All experiments were carried out in strict accordance with the recommendation in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the IACUC of Rush University and University of Illinois at Chicago Committee on Animal Resources.
Butyrate treatment in G93A mice
G93A 8–9 weeks old mice were divided into two groups randomly. The butyrate-treated group receives sodium butyrate (Sigma-Aldrich, 303410) at a 2% concentration in filtered drinking water. Control group receives filtered drinking water without sodium butyrate. All mice are provided with water ad libitum, and maintained in a 12 h dark/light cycle. All animals are weighted and received detailed clinical examination, which included appearance, movement and behavior patterns, skin and hair conditions, eyes and mucous membranes, respiration and excreta. If restricted outstretching of the hind legs is observed when the tail is hold, it means the symptom of ALS is obvious. Lie the mouse on the back, if it can’t turn over in 30 seconds, the mouse is humanely sacrificed.
Western blot analysis and antibodies
Mouse intestinal mucosal cells were collected by scraping from mouse colon, including proximal and distal colon, as previously described. 10 Briefly, mouse mucosal cells were lysed in lysis buffer (1% Triton X-100 (Sigma-Aldrich, X100), 150 mM NaCl (J.T. Baker 3624-19), 10 mM Tris (Fisher Scientific, BP152-5) pH 7.4, 1 mM EDTA (Fisher Scientific, BP120-1), 1 mM EGTA (Sigma-Aldrich, E3889) pH 8.0, 0.2 mM sodium ortho-vanadate (Sigma-Aldrich, S6508) and protease inhibitor cocktail (Roche Diagnostics, 118367001). Cultured cells were rinsed twice in ice-cold Hanks’ balanced salt solution (Sigma-Aldrich, H1387), lysed in protein loading buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol (Amresco, 0281), 2% SDS (Sigma-Aldrich, L3771), 0.1% bromophenol blue (IBI Scientific, IB74040) and 10% glycerol (Sigma-Aldrich, G5516)) and sonicated (Branson Sonifier, 250). Equal amount of protein was separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose (Bio-rad, 162-0112) and immunoblotted with primary antibodies: Villin (Santa Cruz, sc7672), ZO-1 (Invitrogen, 33–9100) or β-actin (Sigma-Aldrich, A1978) antibodies and visualized by ECL chemiluminescence (Thermo Scientific, 32106). Membranes probed with more than one antibody were stripped before re-probing. Western blot bands were quantified using Image Lab 4.01 (Bio-Rad).
Immunofluorescence
Intestinal tissues were freshly isolated and paraffin-embedded after fixation with 10% neutral buffered formalin. Immunofluorescence was performed on paraffin-embedded sections (5 µm). After preparation of the slides as described previously10, tissue samples were incubated with anti-lysozyme (Santa Cruz, sc27958), ZO-1, or human SOD-17 at 4°C overnight. Samples were then incubated with sheep anti-goat Alexa Fluor 594 (Life Technologies, A11058), or goat anti-mouse Alexa Flour 488 (Life Technologies, A-11001) and DAPI (Life Technologies, D1306) for 1 h at room temperature. Tissues were mounted with SlowFade (Life Technologies, s2828), followed by a coverslip, and the edges were sealed to prevent drying. Specimens were examined with Zeiss laser scanning microscope (LSM) 710.
Fluorescence Permeability in vivo
Fluorescein Dextran (Molecular weight 3000 Da, diluted in Hanks) was gavaged (50 mg/g mouse). Four hours later, mouse blood samples were collected by cardiac puncture. Fluorescence intensity of the plasma was measured on a fluorescent plate reader, as previously described. 11, 12
Paneth cells counting
Paneth cells in mouse ileal cells were counted after anti-lysozyme immunofluorescence staining. The patterns of lysozyme expression in Paneth cells were classified into two categories: normal and abnormal which included disordered, depleted and diffuse according to published methods. 10, 13
HCT116 cell transfection and live cell imaging
The HCT116 cells were first transfected with the plasmids for fluorescent protein tagged human wild type SOD1-GFP or human ALS-causing mutation SOD1G93A-GFP. The HCT116 cells with SOD1G93A-GFP transfection were cultured with or without 2 mM Butyrate. Forty-eight hours after transfection, the live HCT116 cells were used for evaluating the protein aggregate formation under a confocal microscope (Leica SP8). Three independent sets of experiments were conducted. With GFP expression, the intracellular protein aggregates can be easily distinguished from the homogenous cytosolic expression. The percentage of cells with protein aggregates was counted in each experimental group.
Real-time quantitative PCR
Total RNA was extracted from mouse ileal epithelial cells or cultured cells using TRIzol reagent (Life Technologies, 15596-02). RNA was first reverse-transcribed into cDNA with the iScript cDNA synthesis kit (Bio-Rad, 170–8891) according to the manufacturer’s manual. The RT-cDNA reaction products were subjected to quantitative real-time PCR using CTFX 96 Real-time system (Bio-Rad, C1000) and SYBR green supermix (Bio-Rad, 172–5124) according to the manufacturer’s directions. All expression levels were normalized to β-actin levels of the same sample. Percent expression was calculated as the ratio of the normalized value of each sample to that of the corresponding untreated control cells. All real-time PCR reactions were performed in triplicate. Primers were designed as described.10
Real-time PCR measurement of bacterial DNA
DNA was extracted as previously described10. 16S rDNA PCR reactions were performed with the following primers: Universal bacteria (forward: 5’-TCCTACGGGAGGCAGCA GT-3’; reverse: 5’-GGACTACCAGGGTATCTAATCCTGTT-3’), Butyrivibrio Fibrisolvens (forward: 5’-CTAACACATGCAAGTCGAACG-3’; reverse: 5’-CCGTGTCTCAGTCCCAAT G-3’), Firmicuyes Peptostreptococus (forward: 5’-CATTGGGACTGAGACAC-3’; reverse: 5’-AATCCGGATAACGCTTGC-3’), Butyryl-CoA transferase (forward: 5’-GCIGAICATTTCACITGGAAYWSITGGCAYATG-3’; reverse 5’-CCTGCCTTTGCAATRTCIACRAANGC-3’) 14. The relative amount of 16S rDNA in each sample was estimated using the ΔΔCT.
Microbial sampling and sequencing
The tubes for microbial sampling were autoclaved and then irradiated with ultraviolet light to destroy the contaminating environmental bacterial DNA. The mice were then anesthetized and dissected. Fresh fecal and cecal stools were isolated from the gut and placed into the specially prepared tubes. The samples were kept at low temperature with dry ice and mailed to the Research and Testing Laboratory (Lubbock, Texas) for 454 pyrosequencing. The sequences were denoised and subjected to quality checks.
The V4-V6 region of the samples was amplified for pyrosequencing in the Research and Testing Laboratory (Lubbock, TX) using forward and reverse fusion primers. The forward primer was constructed (5’-3’) with the Roche A linker (CCATCTCATCCCTGCGTGTCTCCGACTCAG), an 8–10 bp barcode, and the 530F–GTGCCAGCMGCNGCGG primer. The reverse fusion primer was constructed (5’-3’) with a biotin molecule, the Roche B linker (CCTATCCCCTGTGTGCCTTGGCAGTCTCAG), and the 1100R-GGGTTNCGNTCGTTR primer. Amplifications were performed in 25 µl reaction volumes with Qiagen HotStar Taq master mix (Qiagen Inc., Valencia, California), 1 µl of each primer (5 µM), and 1 µl of template. Reactions were performed on ABI Veriti thermocyclers (Applied Biosytems, Carlsbad, California) under the following thermal profile: 95 °C for 5 min, then 35 cycles of 94 °C for 30 sec, 54 °C for 40 sec, 72 °C for 1 min, followed by one cycle of 72 °C for 10 min and a 4 °C hold. The amplicons were 570 bp in length.
Amplification products were visualized with eGels (Life Technologies, Grand Island, New York). Products were then pooled to equimolar levels, and each pool was cleaned with Diffinity RapidTip (Diffinity Genomics, West Henrietta, New York) and size-selected using Agencourt AMPure XP (BeckmanCoulter, Indianapolis, Indiana) following the Roche 454 protocols (454 Life Sciences, Branford, Connecticut). Size-selected pools were then quantified, and 150 ng of DNA was hybridized to Dynabeads M-270 (Life Technologies) to create single-stranded DNA following Roche 454 protocols (454 Life Sciences). Single-stranded DNA was diluted and used in emPCR reactions, which were performed and subsequently enriched. Sequencing followed established manufacturer protocols (454 Life Sciences).
Bioinformatic analysis
16S rRNA detection, clustering and identification
Clustering of the reads with 4% divergence on the seed sequences was performed using USEARCH to identify similar clusters.15 Chimera checking was detected using the de novo method of UCHIIME.16 The sequences were then denoised according to a size criterion. For taxonomic identification, the sequences were clustered into OTUs with 0% divergence with USEARCH.15 The seed sequence was then queried against NCBI using a distributed .NET algorithm, which makes use of BLASTN+ (KrakenBLAST, www.krakenblast.com).
Taxonomic classification
Taxonomic identifications were assigned by queries against NCBI. Imputation of bacterial genomes was performed according to the similarity of sequenced genomes to reference GreenGenes sequences.17 Principal coordinates analysis (PCoA) of unweighted UniFrac distances plots were plotted using Quantitative Insights Into Microbial Ecology (QIIME)(http://qiime.sourceforge.net/).18
Statistical analysis
The data were expressed as the mean±SD. Differences between two samples were analyzed by Student’s t-test. Differences among three or more groups were analysed using ANOVA with GraphPad Prism 5. p Values of 0.05 or less were considered statistically significant.
Results
Feeding the ALS mice with butyrate delayed the ALS progression
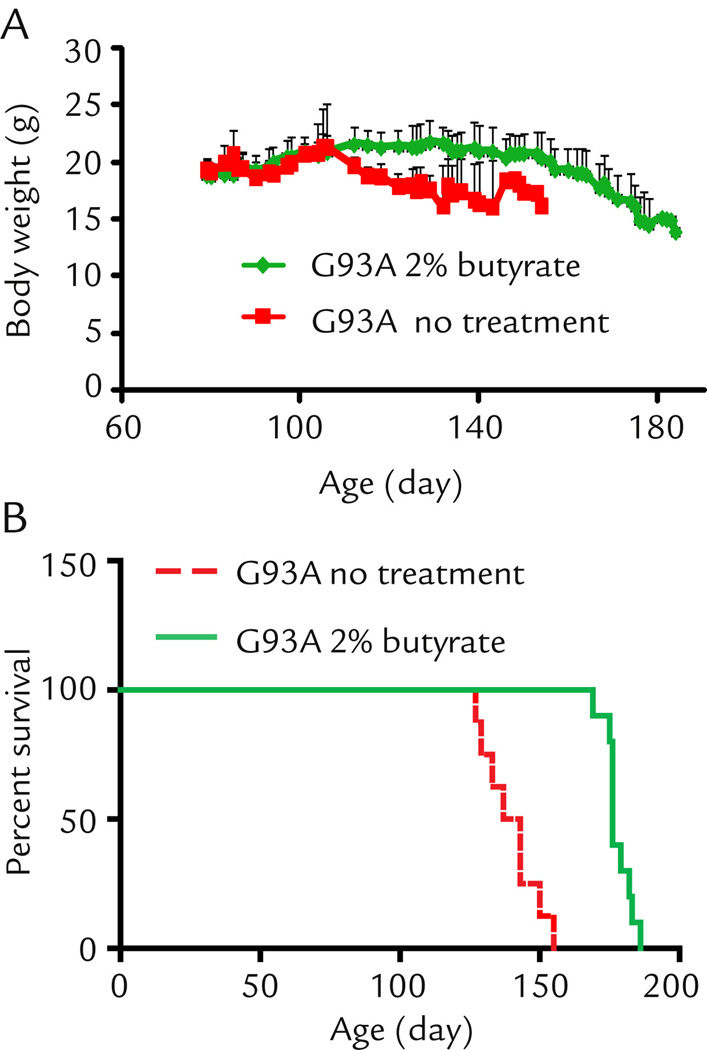
There are early changes of intestinal microbiome before onset of ALS.6 In the current study, we developed a strategy to feed the ALS mice (starting at the age of 1 month old) with a natural bacterial product butyrate (2% butyrate in drinking water). As shown in Fig. 1A, control G93A mice showed weight loss (indication of the disease onset) at the age of 110 days, while the butyrate treated G93A mice did not show weight loss until 150 days. Remarkably, feeding the ALS model mice with butyrate significantly prolonged the life span by 38 days on average (Fig. 1B). These data show that butyrate treatment can slow down ALS disease progression and prolong the life span of G93A mice.
Figure 1. Sodium butyrate prevents body weight loss (A) and extends survival time (B) in G93A amyotrophic lateral sclerosis model mice.
G93A mice were divided into a without treatment group (n=8) and a butyrate group (n=10). Butyrate mice started to drink 2% sodium butyrate in the drinking water bottle from age 63 days, without treatment mice drank regular water mice. Without treatment mice died on day 127, 129, 133, 137, 143 (n=2), 150, 155 separately. With treatment mice died on day 169, 175,176 (n=4), 179, 182, 183, 186 separately.
Reduced population of butyrate-production bacteria and less butyryl-coenzyme A in ALS mice before the onset of diseases
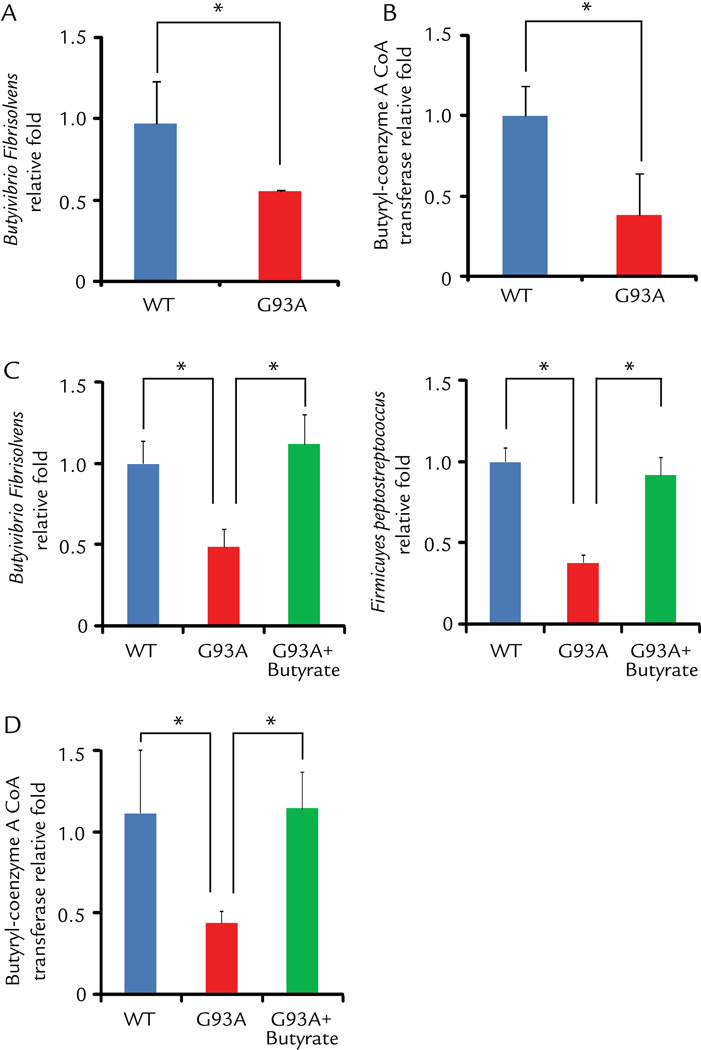
We found that ALS G93A mice showed abnormal intestinal microbiome, in which butyrate-producing bacteria (Butyrivibrio Fibrisolvens)19 were reduced (Fig. 2A). This change occurs in young ALS mice before disease onset. Butyrate synthesis by anaerobic bacteria can occur via butyryl-coenzyme A (CoA):acetate CoA-transferase.20 We also found that the level of CoA enzyme was significantly lower in the G93A mice without disease symptom, compared to those in the wild-type (WT) and transgenic G93A mice (Fig. 2B). Strikingly, oral 2% sodium butyrate treatment for 2.5 months significantly enhanced the abundance of butyrate-producing bacteria Butyrivibrio Fibrisolvens (Fig. 2C). Firmicutes make up the largest portion of the mouse and human gut microbiome. 21 The division Firmicutes as part of the gut flora has been shown to be involved in energy resorption and obesity. 22–24 Fimicutes Peptostreptococus, which was low in ALS mice,6 was also enhanced after butyrate treatment (Fig. 2C). We found that butyrate treatment significantly restored the expression of the level of CoA enzyme (Fig. 2D).
Figure 2. Changes of butyrate-producing bacteria and butyryl-coenzyme A CoA transferase in the intestine of ALS mice treated by butyrate.
(A) Butyrate-producing bacteria Butyrivibrio Fibrisolvens decreased in G93A ALS model mice (2 month old, without symptom). (B) Expression of Butyryl-coenzyme A CoA transferase gene decreased in G93A amyotrophic lateral sclerosis model mice. (C)Butyrivibrio Fibrisolvens and Fimicuyes peptostreptococus enhanced in ALS mice treated with 2% sodium butyrate for 2.5 months. (D) Butyryl-coenzyme A CoA transferase genes enhanced in ALS mice with butyrate treatment. Primers specific to universal 16s rRNA were used as an endogenous control to normalize loading between samples. The relative amount of each sample was estimated using the ΔΔCT method. (n=3, Student’s t-test. *P<0.05).
Butyrate treatment corrected dysbiosis in ALS mice
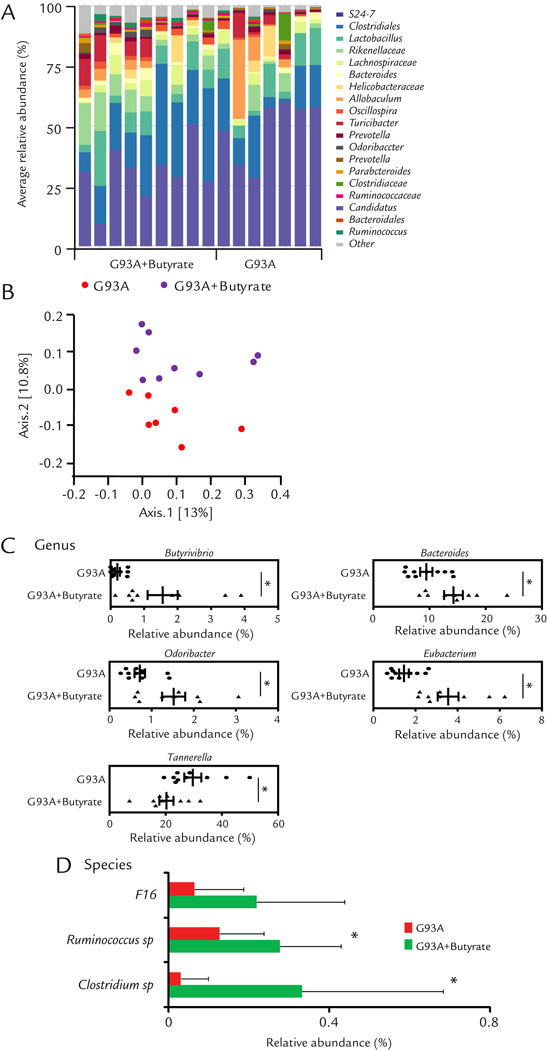
We then collected the fecal samples of ALS mice with or without butyrate for the 16S rDNA sequencing. We identified changes of gut microbial profile in ALS mice with butyrate treatment (Fig. 3A). The microbiome of ALS mice and butyrate treated ALS mice fell into different clusters on a PCoA scale (Fig. 3B). Specifically, we analyzed the OTU table generated by sequencing to identify the bacterial changes in taxonomic assignment. Genus level changes were analyzed to evaluate the effects of butyrate treatment on the taxonomic assignment of intestinal microbiota (Fig. 3C). We checked butyrate-producing bacteria (Butyrivibrio) and then found butyrate treatment significantly increased Butyrivibrio at genus level. This results further confirmed our previous data derived from RT-PCR. Butyrate treatment was able to significantly increase the abundance of other butyrate-producing bacteria including Bacteroide25, Odoribacter25 and Eubacterium19, 26. Interesting, Butyrate treatment also significantly depleted Tannerella27, which induced foam cell formation in THP-1 cells and accelerated the progression of atherosclerotic lesions.
Figure 3. Dysbiosis restored by the butyrate treatment in ALS mice.
(A) Representative bacterial community of fecal samples from G93A and butyrate treatment mice, using 454 16S rRNA sequencing data. (B) Representative Principal Coordinates Analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes (G93A+Butyate group:n = 9 and G93A group:n = 10). (C) and (D) Alterations of taxonomic assignment of the intestinal bacteria with butyrate treatment at genus and species levels. Butyrate treatment changed microbial profile in intestine of ALS mice (G93A+Butyate group: n = 9 and G93A group: n = 10, student’s t-test *P<0.05.)
At the species level, at the age of 3.5 months (1 month old G93A mice after butyrate treatment for 2.5 months), the ALS mice with butyrate treatment, significantly enriched Clostridium sp. and Ruminococcus sp (Fig. 3D). Taken together, these data indicate that Butyrate treatment corrected dysbiosis and enhanced diversity of microbiome in ALS mice.
Correct abnormal intestinal tight junction structure in ALS mice with butyrate treatment
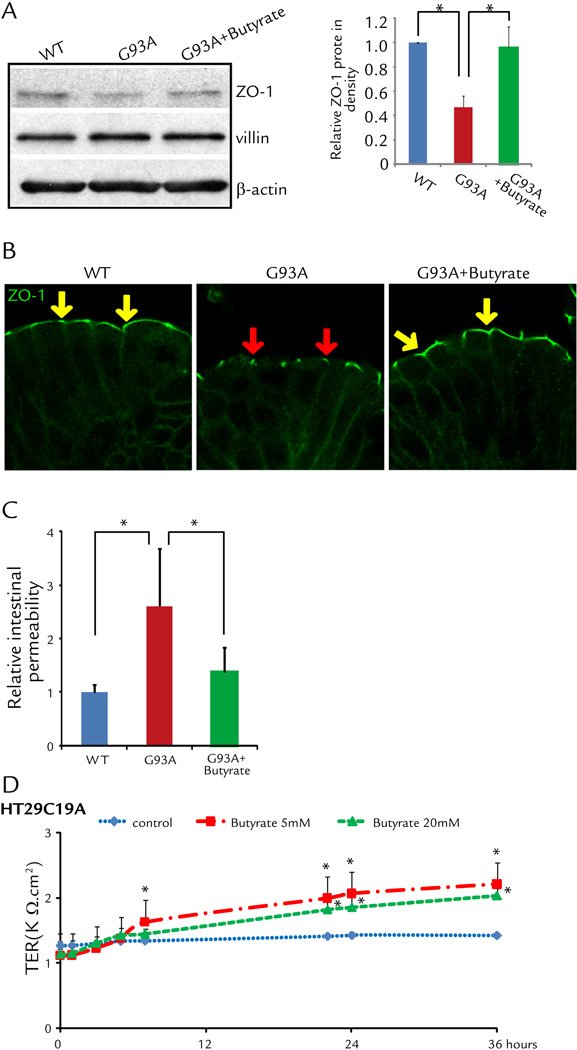
The intestinal barrier is maintained by tight junctions (TJ) formed between adjacent intestinal epithelial cells. We have discovered early changes of intestinal TJs in the G93A mice before onset of ALS.6 We further hypothesize that restoring intestinal homeostasis delays ALS disease onset and progression. We found that butyrate treatment for 2.5 months (started at the age of 1 month) significantly restored the expression of ZO-1 (a TJ-protein) in the G93A gut (Fig. 4A). We performed immunostaining of ZO-1 and observed the zipper-like tight junction structure in the intestine of WT mice (yellow arrows), whereas TJ structure was damaged in the G93A intestinal epithelial cells (red arrows) (Fig. 4B). Butyrate treatment improved the TJ structure in the G93A gut (yellow arrows) (Fig. 4B-right panel). These data indicate butyrate treatment was able to correct abnormal intestinal tight junction structure in ALS mice.
Figure 4. Correct abnormal intestinal tight junction structure in ALS mice with butyrate treatment.
(A) Western blots of ZO-1 in the colon. The relative band intensities of ZO-1 is presented as the means ± SD (n = 5 per group, student’s t-test, *P < 0.05). Western blot bands were quantified using Image Lab 4.01 (Bio-Rad). G93A mouse treated with 2% sodium butyrate for 2.5 months. (B) Immunostaining of ZO-1 in the colon. Data are from a single experiment and are representative of n = 5 mice per group. Arrows show the staining of ZO-1 protein on the top of the intestinal crypts. G93A mouse treated with 2% sodium butyrate for 2.5 months. (C) Intestinal permeability was decreased in ALS mice with butyrate treatment (n = 5, student’s t-test, *P < 0.05). (D) Human epithelial HT29C19A cells increased TER after butyrate treatment. (n = 3, ANOVA test, *P < 0.05).
Correct leaky gut in ALS mice with butyrate treatment
The intestinal permeability was assayed by gavaging mice with Fluorescein Dextran (50 mg/g mouse). Four hours later, mouse blood samples were collected for fluorescence intensity of the plasma. The serum level of fluorescein is about 3-fold higher in G93A mice (Fig. 4C), indicating that G93A mice have gut leakage. The butyrate treatment significantly decreased intestinal permeability in the ALS mice (Fig. 4C). To examine the role of butyrate in altering intestinal permeability, we used a polarized human intestinal epithelial cell model in vitro. Our data further showed human HT29C19A cells had significantly higher trans-epithelial resistant (TER) after butyrate treatment, indicating the role of butyrate in correcting leaky gut through enhancing intestinal epithelial function (Fig. 4D).
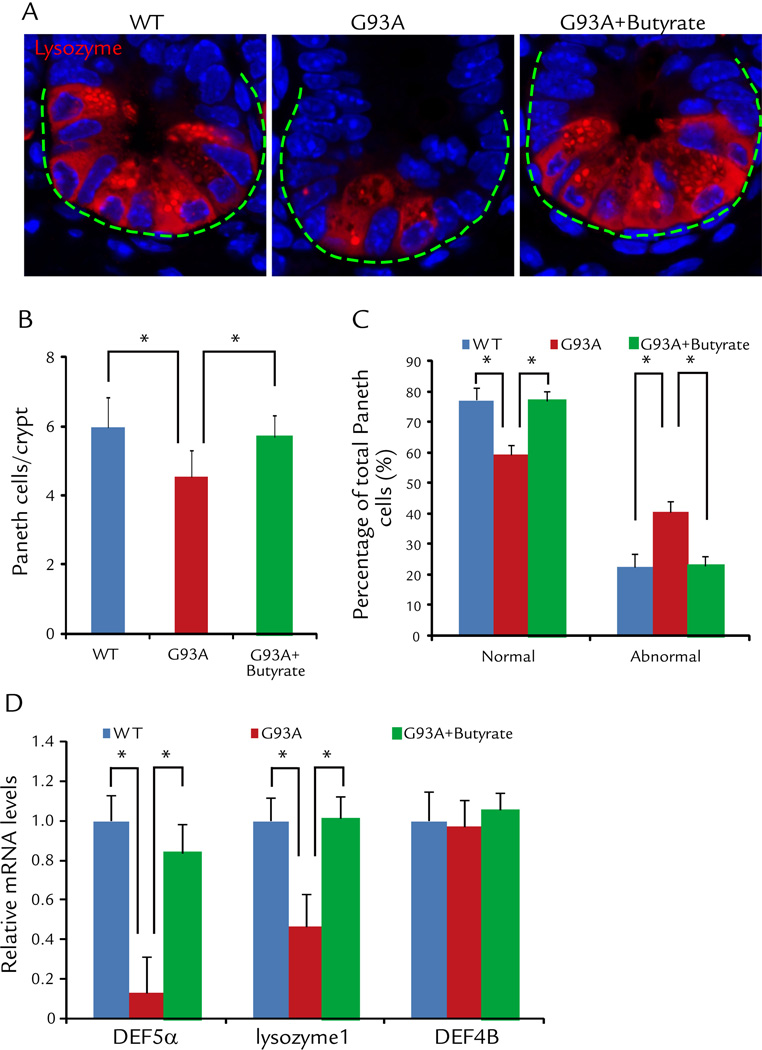
Abnormal Paneth cells were decreased in the ALS mice treated by butyrate
Paneth cells are specialized intestinal epithelial cells associated with autophagy activity and regulate the host-bacterial interactions in gut.10, 13 We specifically notice the decreased normal Paneth cells (Fig. 5A), shown as number of Paneth cell per crypt (Fig. 5B). The percentage of abnormal Paneth cells was also significantly decreased in intestinal of ALS mice treated with butyrate, compared with the non-treated mice (Fig. 5C). The lysozyme 1 and anti-microbial peptide defensin 5 alpha was also restored in the ALS intestine after butyrate treatment (Fig. 5D). These data indicate the restoration of Paneth cells in intestine of ALS mice with butyrate treatment.
Figure 5. Butyrate corrects defects in Paneth cells in intestine of ALS mice treated with butyrate.
(A) Immunostaining of lysozyme in the small intestine of ALS mice treated with butyrate. (B) Number of Paneth cell/crypt was significantly increased in intestinal of ALS mice treated with butyrate, compared with the non-treated mice. (n = 5, student’s t-test, *P < 0.05). (C) The percentage of abnormal Paneth cells was significantly decreased in intestinal of ALS mice treated with butyrate, compared with the non-treated mice. (n = 5, student’s t-test, *P < 0.05). (D) Lysozyme 1 and anti-microbial peptide defensin 5 alpha were restored in the ALS intestine after butyrate treatment. (n = 3, student’s t-test, *P < 0.05).
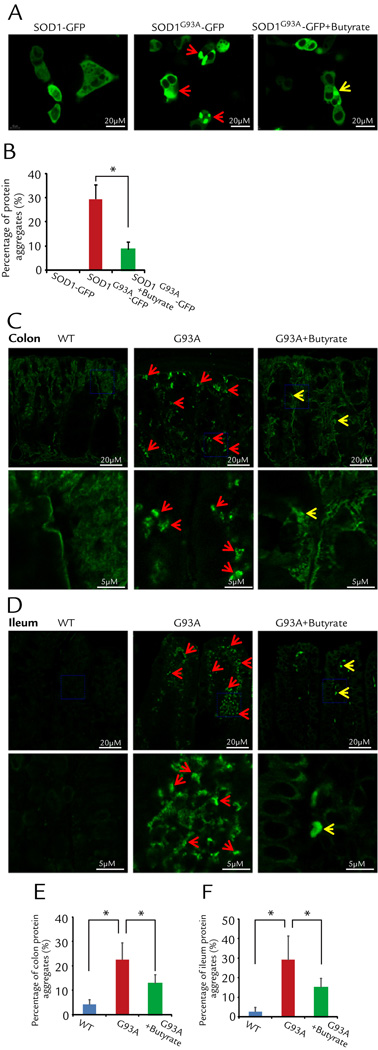
Butyrate treatment decreased aggregation of SOD1 mutant protein in human intestinal epithelial cells
Aggregation of SOD1 mutant protein in motor neurons and skeletal muscle is a hallmark of ALS pathology.7–9 We first tested whether SOD1G93A also forms aggregates in the intestinal epithelial cells. The human intestinal epithelial cells (HCT116) were transfected with fluorescent protein tagged human wild type SOD1-GFP or human ALS mutation SOD1G93A-GFP. Remarkably, only SOD1G93A forms protein aggregates inside the intestine epithelial cells, while not a single cell transfected expressing wild type SOD1-GFP shows protein aggregates. The representative images are shown in Fig. 6A. We further examined whether butyrate treatment could reduce the protein aggregation of SOD1G93A in the HCT116 cells. Two groups of HCT116 cells were transfected with SOD1G93A-GFP, twenty-four hours later, the group with 2 mM butyrate in the culture medium showed significant reduction in protein aggregate formation (Fig. 6B).
Figure 6. Butyrate treatment decreased aggregation of SOD1 mutant protein in human intestinal epithelial HCT116 cells and intestine of ALS mice.
(A) Representative images from three independent experiments for HCT116 cells transfected with SOD1-GFP or SOD1G93A-GFP 48 hours after the transfection. The wild type SOD1-GFP shows a pattern of a homogenous cytosolic expression. Not a single cell expressing wild type SOD1-GFP shows protein aggregates. Clearly, the cells with SOD1G93A-GFP expression show cytosolic protein aggregates (white arrows). Remarkably, application of 2 mM butyrate in the culture medium reduced the protein aggregation in the HCT116 cells expressing SOD1G93A-GFP. (B) Summary of the three independent experiments: 30.1% ± 2.7% of SOD1G93A-GFP cells show protein aggregates, while only 8.7% ± 1.2% SOD1G93A-GFP cells show protein aggregates in the present of 2mM butyrate (n=3, student’s t-test,* P<0.05). (C) and (D) ALS mice have aggregation of SOD1 mutant protein and butyrate treatment decreased aggregation of SOD1 mutant protein in colon (C) and ileum (D) in vivo. Arrows indicate the aggregation of SOD1 mutant protein. The high magnitude images showed the details from the blue frames in colon and ileum. (E) and (F) Summary of aggregation of SOD1 mutant protein in colon and ileum with 2% butyrate treatment in drinking water for 2.5 months (n=5, student’s t-test,* P<0.05).
In vivo, we found enhanced protein aggregation of SOD1G93A in colon of the G93A mice, compared to that of WT mice (Fig. 6C and D). Butyrate treatment could significantly reduce the SOD1G93A aggregation in the intestine of ALS mice (Fig. 6E and F).
Discussion
In the current study, we reported that aberrant microbiome and lost intestinal homeostasis is associated with ALS pathophysiology in the mouse model. Restoring microbiome and intestinal homeostasis using bacterial product butyrate preserves neuromuscular function, thus delaying disease onset and progression. Our study provides the first evidence of gut dysfunction and microbiome as an important component in ALS disease progression. Most importantly, our study opens a new door to develop novel strategies to attenuate the phenotype of ALS and delay the disease.
Butyrate is produced by microbes through fermentation in the colon. It is considered a major source of energy for colonocytes through assimilation in TCA cycle, and as an HDAC (histone deacetylase) inhibitor28. Donohoe et al. 29 showed that the function of butyrate goes beyond simple metabolism. Butyrate could rescue deficit in mitochondrial respiration and diminished oxidative metabolism in germ-free colonocytes. The mechanism is due to butyrate acting as an energy source rather than as an HDAC inhibitor. In fact, butyrate enema therapy has been shown to restore intestinal homeostasis and ameliorate the inflammation associated with colitis in mouse models and in human clinical trials.30, 31 Butyrate belongs to short chain fatty acids (SCFA), which modulates the physiology of the host through binding G protein-coupled receptors.32 In addition to its local beneficial functions in intestine, butyrate enters the bloodstream via colonic absorption.33–35 Adiposity,32 T cell differentiation,36 and inflammatory responses37 are modulated by SCFAs. It is possible that increasing butyrate or other SCFA levels help reverse pathophysiological changes in various human diseases, including ALS.
Our study with oral administration of butyrate as a natural bacterial product to effectively restore microbial and intestinal homeostasis presents a different and better therapeutic strategy, as evidenced by the prolonged life span by 38 days on average in the butyrate treated ALS mice. As a pharmacological reagent and a butyrate derivative, phenylbutyrate (400mg/kg), extends the survival of G93A mice for 29 days.38 Phenylbutyrate has been in Phase 2 clinical trial for ALS.39 Phenylbutyrate was safe and tolerable when it was used for 20 weeks under the dosage of 9–21g/day in ALS patients. However, the original study in G93A mice showed that 400 mg/kg/day was the optimal dosage to obtain the maximum 29 days extension of survival, while a little higher dosage is toxic to the mice. G93A mice treated with 600–800mg/kg/day of phenylbutyrate became ill and died earlier than untreated G93A mice.38 Phenylbutyrate has been tested in ALS mice through intraperitoneal injection in previous reports under the mechanism of inhibition of histone deacetylase activity (HDAC).38 It is likely that the primary effect of oral butyrate application is to improve the gut defects in ALS mice rather than via HDAC inhibition.
The exact role of butyrate on intestinal and microbial function in ALS has not been examined previously. It is very well known that the ALS-causing SOD1 mutation leads to protein aggregate formation in neuron and skeletal muscle cells.40 However, it has never been tested whether the ALS-causing SOD1 mutation causes similar pathological phenotype in the intestine. We, for the first time, demonstrated that SOD1G93A mutant protein indeed forms aggregates in the intestine of ALS mice and human intestinal epithelial cells, suggesting that SOD1G93A plays an essential role in pathophysiological alterations in the intestine. Moreover, the percentage of abnormal Paneth cells was significantly decreased in the intestine of ALS mice treated with butyrate. The lysozyme 1 and anti-microbial peptide defensin 5 alpha was also restored in the ALS intestine after butyrate treatment. The pathological changes likely mirrors population shifts in the microbiome in G93A mice. Remarkably, the dysfunction of microbiome and intestinal homeostasis in the G93A mice were restored by oral application of butyrate.
We recognize that butyrate treatment is not a cure of the genetic defect in ALS G93A mice, rather to improve and slow down the pathological progression in the intestine that plays an important role in maintaining physiological integrity of the whole body. In addition, current literature supports the role of butyrate in regulating metabolic function of mitochondria. It is possible that butyrate improves the mitochondrial function in general in ALS mice.29, 41–44 Thus, the treatment delays the degeneration of neuromuscular system until the genetic defect overrides the beneficial role of butyrate. Butyrate is a hydrophobic molecule and may be orally absorbed and distributed systemically. It is an energy substrate within various cells48. Thus, butyrate may have a direct effect on neuron functions. Our current study highlights the complex role of gut microbiome and intestinal epithelium in the progression of ALS and presents butyrate as a potential therapeutic reagent to restore ALS dysbiosis. Understanding the detailed molecular mechanism underlying the direct beneficial effects of butyrate and gut microbiome on the neuromuscular system is beyond the scope of the current study, which is an undergoing study in our research team.
The early GI problems in ALS patients before they show neurodegenerative symptoms have been largely ignored. It is reported that lipopolysaccharides (LPS) was increased in the serum of ALS patients.45 In ALS patients, constipation is common and presumed multifactorial - related to dehydration, lack of dietary fiber intake, and decreased physical activity.46 However, we lack research on the early GI issue from ALS patients. ALS is a systemic disorder that involves dysfunction of multiple organs.47 We believe that ALS-associated mutations not only cause defects in neuron, muscle and skeletal systems, but also in intestine and microbiome. A key question is whether and how the intestinal integrity and microbiome is altered in ALS patients before the onset of symptoms. ALS patients do not go to the clinic until the symptoms occur. It is hard to collect intestinal biopsies from ALS patients. Thus, further mechanistic studies need to be conducted using ALS animal models.
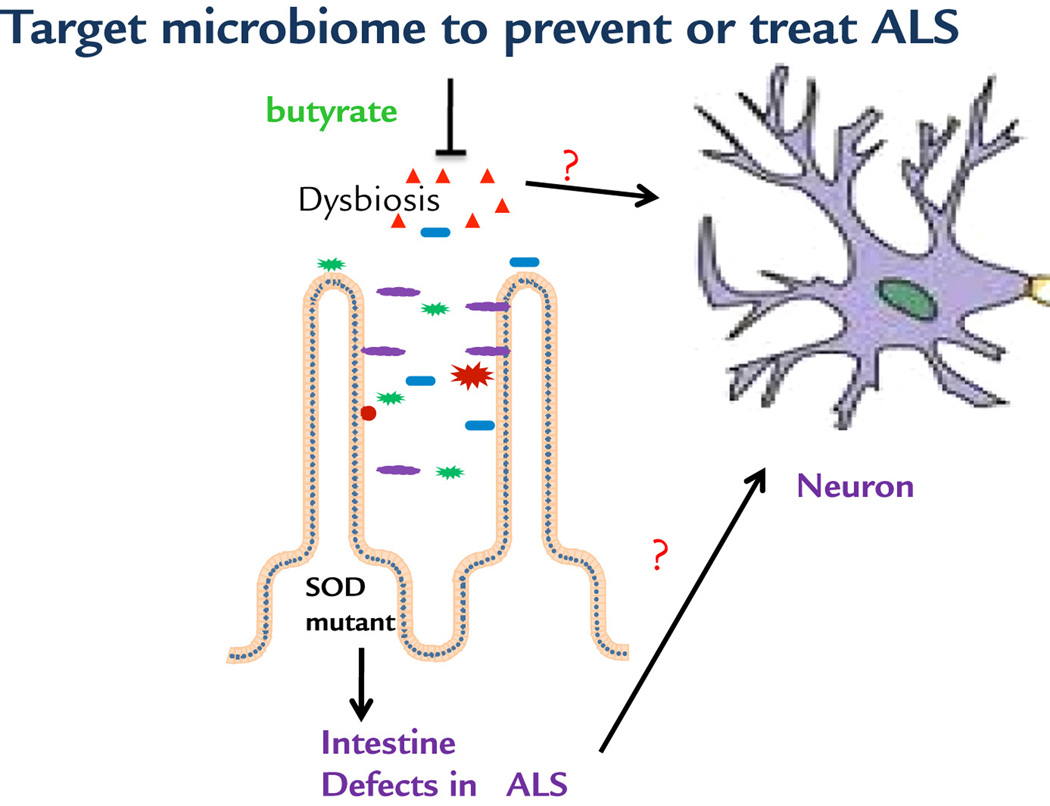
Our observations allow us to propose a working model of microbial and intestinal homeostasis that may contribute to the neuromuscular dysfunction in ALS (Fig. 7). ALS-linked SOD1G93A mutation causes intestinal dysfunction and aberrant microbiome as part of the ALS pathology. Targeting the crosstalk between gut and neuromuscular systems corrects the dysbiosis and restores the intestinal tight junctions, Paneth cells, and beneficial bacteria, thus prolonging the life span of ALS mice. It represents a novel means for preserving the integrative physiology in ALS. Our findings demonstrated that gut microbiome may play essential roles in the pathogenesis and pathophysiology of diseases in distant organs. It brings novel concepts to the ALS research field and has significant translational implications for identifying new biomarkers for ALS diagnosis and developing new therapeutic strategies to combat this devastating disease.
Fig 7. A working model of microbial and intestinal homeostasis that may contribute to the neuromuscular dysfunction in ALS.
Butyrate treatment corrects the intestinal dysfunction and gut dysbiosis, thus improving neuron and muscle function. It is unknown whether the beneficial role of butyrate is directly or indirectly on neuron and muscle function.
Acknowledgments
We would like to acknowledge the NIDDK/National Institutes of Health grant R01 DK105118 to JS, NIAMS R01 AR057404 to JZ, and ALS Association Research Award 2015 to JZ and JS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interests:
The authors have indicated that they have no other conflicts of interest regarding the content of this article.
References
- 1.Alonso A, Logroscino G, Jick SS, Hernan MA. Incidence and lifetime risk of motor neuron disease in the United Kingdom: a population-based study. European journal of neurology : the official journal of the European Federation of Neurological Societies. 2009;16:745–751. doi: 10.1111/j.1468-1331.2009.02586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joyce PI, Fratta P, Fisher EM, Acevedo-Arozena A. SOD1 and TDP-43 animal models of amyotrophic lateral sclerosis: recent advances in understanding disease toward the development of clinical treatments. Mammalian genome : official journal of the International Mammalian Genome Society. 2011;22:420–448. doi: 10.1007/s00335-011-9339-1. [DOI] [PubMed] [Google Scholar]
- 3.Forsyth CB, Shannon KM, Kordower JH, et al. Increased intestinal permeability correlates with sigmoid mucosa alpha-synuclein staining and endotoxin exposure markers in early Parkinson’s disease. PloS one. 2011;6:e28032. doi: 10.1371/journal.pone.0028032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurney ME, Pu H, Chiu AY, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 6.Wu S, Yi J, Zhang YG, Zhou J, Sun J. Leaky intestine and impaired microbiome in an amyotrophic lateral sclerosis mouse model. Physiol Rep. 2015:3. doi: 10.14814/phy2.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng HX, Shi Y, Furukawa Y, et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanova MI, Sievers SA, Guenther EL, et al. Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:197–201. doi: 10.1073/pnas.1320786110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Lee H, Lee JH, et al. Dimerization, oligomerization, and aggregation of human Amyotrophic lateral sclerosis Cu/Zn-superoxide dismutase 1 mutant forms in live cells. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M113.542613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S, Zhang YG, Lu R, et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2014 doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YG, Wu S, Xia Y, Sun J. Salmonella infection upregulates the leaky protein claudin-2 in intestinal epithelial cells. PloS one. 2013;8:e58606. doi: 10.1371/journal.pone.0058606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao AP, Petrof EO, Kuppireddi S, et al. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PloS one. 2008;3:e2369. doi: 10.1371/journal.pone.0002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Applied and environmental microbiology. 2007;73:2009–2012. doi: 10.1128/AEM.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 16.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHardy IH, Li X, Tong M, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS microbiology letters. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 20.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Applied and environmental microbiology. 2002;68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 23.Henig RM. Fat Factors. New York Times Magazine. 2008 Sep 28; [Google Scholar]
- 24.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. 2014;5:e00889. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barcenilla A, Pryde SE, Martin JC, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Applied and environmental microbiology. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HR, Jun HK, Choi BK. Tannerella forsythia BspA increases the risk factors for atherosclerosis in ApoE(−/−) mice. Oral diseases. 2014;20:803–808. doi: 10.1111/odi.12214. [DOI] [PubMed] [Google Scholar]
- 28.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 29.Donohoe DR, Garge N, Zhang X, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamer HM, Jonkers DM, Bast A, et al. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clin Nutr. 2009;28:88–93. doi: 10.1016/j.clnu.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 32.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 37.Le Poul E, Loison C, Struyf S, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. The Journal of biological chemistry. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 38.Ryu H, Smith K, Camelo SI, et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. Journal of neurochemistry. 2005;93:1087–1098. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- 39.Cudkowicz ME, Andres PL, Macdonald SA, et al. Phase 2 study of sodium phenylbutyrate in ALS. Amyotroph Lateral Scler. 2009;10:99–106. doi: 10.1080/17482960802320487. [DOI] [PubMed] [Google Scholar]
- 40.Redler RL, Dokholyan NV. The complex molecular biology of amyotrophic lateral sclerosis (ALS) Progress in molecular biology and translational Science. 2012;107:215–262. doi: 10.1016/B978-0-12-385883-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Z, Yin J, Zhang J, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolar S, Barhoumi R, Jones CK, et al. Interactive effects of fatty acid and butyrate-induced mitochondrial Ca(2)(+) loading and apoptosis in colonocytes. Cancer. 2011;117:5294–5303. doi: 10.1002/cncr.26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y, Chen Y, Jiang H, Nie D. The role of short-chain fatty acids in orchestrating two types of programmed cell death in colon cancer. Autophagy. 2011;7:235–237. doi: 10.4161/auto.7.2.14277. [DOI] [PubMed] [Google Scholar]
- 44.Tang Y, Chen Y, Jiang H, Nie D. Short-chain fatty acids induced autophagy serves as an adaptive strategy for retarding mitochondria-mediated apoptotic cell death. Cell Death Differ. 2011;18:602–618. doi: 10.1038/cdd.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang R, Miller RG, Gascon R, et al. Circulating endotoxin and systemic immune activation in sporadic amyotrophic lateral sclerosis (sALS) Journal of neuroimmunology. 2009;206:121–124. doi: 10.1016/j.jneuroim.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forshew DA, Bromberg MB. A survey of clinicians’ practice in the symptomatic treatment of ALS. Amyotrophic lateral sclerosis and other motor neuron disorders : official publication of the World Federation of Neurology, Research Group on Motor Neuron Diseases. 2003;4:258–263. doi: 10.1080/14660820310017344. [DOI] [PubMed] [Google Scholar]
- 47.Sun J, Zhou J. Does the gut drive ALS progression? Future Medicine. 2015;5:375–378. [Google Scholar]
- 48.Iannitti T, Beniamino Palmieri B. Clinical and experimental applications of sodium phenylbutyrate. Drug R D. 2011;11:227–249. doi: 10.2165/11591280-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]