Abstract
Leprosy is a chronic inflammatory disease caused by Mycobacterium leprae that mainly affects the skin and peripheral nervous system, leading to a high disability rate and social stigma. Previous studies have shown a contribution of genes encoding products of the lectin pathway of complement in the modulation of the susceptibility to leprosy; however, the ficolin-3/FCN3 gene impact on leprosy is currently unknown. The aim of the present study was to investigate if FCN3 polymorphisms (rs532781899: g.1637delC, rs28362807: g.3524_3532insTATTTGGCC and rs4494157: g.4473C>A) and ficolin-3 serum levels play a role in the susceptibility to leprosy. We genotyped up to 190 leprosy patients (being 114 (60%) lepromatous), and up to 245 controls with sequence-specific PCR. We also measured protein levels using ELISA in 61 leprosy and 73 controls. FCN3 polymorphisms were not associated with disease, but ficolin-3 levels were higher in patients with FCN3 *2B1 (CinsA) haplotype (p = 0.032). Median concentration of ficolin-3 was higher in leprosy per se (26034 ng/mL, p = 0.005) and lepromatous patients (28295 ng/mL, p = 0.016) than controls (18231 ng/mL). In addition, high ficolin-3 levels (>33362 ng/mL) were more common in leprosy per se (34.4%) and in lepromatous patients (35.5%) than controls (19.2%; p = 0.045 and p = 0.047, respectively). Our results lead us to suggest that polymorphisms in the FCN3 gene cooperate to increase ficolin-3 concentration and that it might contribute to leprosy susceptibility by favoring M. leprae infection.
Author summary
Leprosy is considered a neglected disease and still a public health problem in many countries where it was not yet eliminated, leading to a high disability rate and social stigma. The molecular mechanisms of M. leprae infection and immune evasion are still poorly known, raising the need for studies that may contribute to a better understanding of leprosy etiology, as well as improvement in diagnosis and treatment. Ficolin-3 is a soluble molecule of the innate immune system that recognizes a wide range of pathogen-associated molecular patterns leading to complement activation and phagocytosis. We observed high concentration of ficolin-3 in leprosy patients, likely caused by polymorphisms present in intronic regions of FCN3 gene, which may contribute to leprosy susceptibility by favoring M. leprae infection. This is the first study addressing FCN3 polymorphisms and ficolin-3 levels in leprosy, indicating it as a good candidate biomarker associated with the host response against M. leprae.
Introduction
Leprosy is a chronic infectious disease caused by Mycobacterium leprae that mainly affects the skin and peripheral nerves [1] and can cause progressive and permanent damage, if untreated. Despite the disease elimination in 119 of the 122 countries where it was considered a public health problem, Brazil still ranked second in the world, behind India and accounts for 92% of leprosy cases in the Americas [2].
Upon exposure to M. leprae, most individuals are intrinsically resistant to infection. Among those who are susceptible, infection may progresses to a wide spectrum of manifestations, with two polar forms: the tuberculoid leprosy and the lepromatous leprosy. Tuberculoid leprosy is characterized by strong cell-mediated immunity, type 1 cytokine profile, low bacillary load and localized lesions. On the other hand, lepromatous leprosy is characterized by low cellular response, type 2 cytokine profile, high bacillary load and disseminated lesions [3]. There is enough evidence to suggest that susceptibility to leprosy and to different clinical manifestations is markedly influenced by host genetic factors [3–6].
Ficolins (Ficolin-1 or M-Ficolin, Ficolin-2 or L-Ficolin and Ficolin-3 or H-ficolin) are soluble molecules of the innate immune system that recognize a wide range of pathogen-associated molecular patterns (PAMPs) [7,8]. Ficolins form complexes with MASPs (MBL-associated serine proteases or MASPs) and activate complement through the lectin pathway, leading to opsonization and phagocytosis of pathogens, and stimulating the production of inflammatory cytokines and nitric oxide [9]. Most active ficolins are composed of four trimeric subunits. Each monomer is formed by an N-terminal region, a collagen-like domain and a fibrinogen-like domain; important in the oligomerization process, in the MASP/phagocyte interaction and in the recognition of specific PAMPs in pathogens, respectively [9].
Ficolins 1, 2 and 3 are encoded by FCN1 and FCN2 genes on 9q34 and FCN3 on 1p36.11, respectively [10]. In previous studies, we demonstrated that FCN2 gene haplotypes associated with normal ficolin-2 levels have a protective effect against leprosy [11] and that FCN1 gene -271DelT, -399A, -542G, -1981A polymorphisms were associated with susceptibility to leprosy [12]. FCN3 comprises eight exons, one of them being an alternative exon (exon 4). Both FCN3 transcripts, with and without exon 4, occur especially in the lung, but also in the liver, heart, kidney, adrenal gland, breast, spleen, thyroid and visceral adipose tissue, but the shorter transcript is less abundant [13]. Exons are highly conserved: although 164 polymorphic noncoding variants are currently listed in Ensembl, all coding DNA variations (including those synonymous) occur at global frequencies below 1%. The g.1637delC variant (rs532781899) in exon 5 is actually the only one reported to be polymorphic at the global scale [14]. It causes a frameshift, leading to premature termination of the translation product. This generates a truncated protein, unable to perform PAMP recognition and complement activation, which may be associated with repetitive infections in some individuals [15–18].
Ficolin-3 has 299 amino acids and is the most abundant ficolin in serum, with a median concentration of ~19500 ng/mL (range 3000–60300 ng/mL) [17,19]. Low ficolin-3 concentration in serum has already been associated with the pathophysiology of sarcoidosis [20], chemotherapy-related infections in children [21], Crohn's disease [22] and heart failure [23]. On the other hand, high ficolin-3 levels were associated with Systemic Lupus Erythematosus [24], ovarian tumors [25] and seem to be a risk factor for shorter graft survival in kidney transplantation [26]. The impact of FCN3 polymorphisms in other diseases has yet to be explored.
In this work, we investigated whether FCN3 polymorphisms and ficolin-3 serum levels play a role in the susceptibility to leprosy and observed an association between high ficolin-3 levels in serum and the disease.
Material and methods
Ethics statement
The study was approved by Human Research Ethics Committee, Health Sciences Sector at Federal University of Parana, Brazil (approval number: 218.104).
Subjects and samples
Study subjects comprised consecutive outpatients from the Hospital de Clínicas, Federal University of Paraná, State Health Department of Paraná, and inpatients from the Sanitary and Dermatologic Hospital of Paraná, both located in Curitiba, southern Brazil. For all 190 patients (38.4% female; 82.3% Euro-Brazilian, 17.7% Afro-Brazilian; average age of 51.5 years, range 18–94), leprosy was diagnosed on the basis of the clinical and histopathological features of affected lesions and classified according to the criteria of Ridley and Jopling [27]. The initial diagnosis was lepromatous leprosy for 114 (60%), tuberculoid leprosy for 15 (7.9%), and borderline leprosy for 28 patients (14.7%); 10 patients (5.3%) had an undetermined form of leprosy and 23 (12.1%) were unspecified. As control subjects, 245, unrelated, symptom-free blood donors from HEMEPAR (Centro de Hematologia e Hemoterapia do Paraná) were assessed (53% female; 80% Euro-Brazilian, 15% Afro-Brazilian; average age of 37.7 years, range 18–61). Patients and control subjects had a similar socioeconomic status, were from the same geographical area, and shared the same ethnic background. All patients and control subjects provided written informed consent.
FCN3 genotyping
DNA extraction was performed using QIAamp DNA extraction kits (Qiagen) according to the manufacturer’s instructions. Three FCN3 SNPs were assessed by sequence-specific amplification method (PCR-SSP), being: g.1637delC (rs532781899) in exon 5; g.3524_3532insTATTTGGCC (rs28362807) in intron 5 and g.4473C>A (rs4494157) in intron 7 (Table 1). Although there are other noncoding polymorphisms not in LD with those selected, they do not tag a haplotype block in the Iberian population (data from the 1000 Genomes project), which is representative for most Euro-Brazilians, as do the intronic polymorphisms chosen for this study. FCN3_Ex5_1637del_R or FCN3_Ex5_1637C_R were conjugated with FCN3_Ex5_F primer to generate a fragment of 748 bp. An amplification control fragment of 500 bp of FCN2 gene was simultaneously generated. FCN3_In5_3524_3532del_F or FCN3_In5_3524_3532ins_F were conjugated with FCN3_In7_4473A_R or FCN3_In7_+4473C_R primer to generate a fragment of 984 bp. A control fragment of 431 bp of HGH gene was simultaneously generated. The intron 5_intron 7 haplotypes were determined without having to infer their phase on the chromosomes due to the PCR-SSP approach with primers annealing on two different SNPs (S1 Fig).
Table 1. Primers used for FCN3 sequence-specific amplification.
| dbSNP | Gene region | Allelesa | 5’-3’ forward primer | 5’-3’ reverse primer | Amplicon | ||
|---|---|---|---|---|---|---|---|
| rs532781899 | Exon 5 | g.1637C | FCN3_Ex5_F | TAGGGTGGGATCTCTGCTTG | FCN3_Ex5_1637C_R | TGTCACAAAAGACTGGGAGGG | 748 bpb |
| g.1637del | FCN3_Ex5_1637del_R | TGTCACAAAAGACTGGGAGGC | |||||
| rs28362807 | Intron 5 | g.3524_3532del | FCN3_In5_3524_3532del_F | GCCACCAAGCGTTCTTGG | 984 bpc | ||
| g.3524_3532ins | FCN3_In5_3524_3532ins_F | CCACCAAGCGTGGCCAAA | |||||
| rs4494157 | Intron 7 | g.4473C | FCN3_In7_4473C_R | GAGGAGGAAACTGAGGCTCAG | |||
| g.4473A | FCN3_In7_4473A_R | GAGGAGGAAACTGAGGCTCAT | |||||
aPosition of polymorphisms are counted with respect to the FCN3 translation start site with the A of ATG being +1
bPCR-SSP endogenous control: 500 bp of the FCN2 gene (forward primer 5’GCCAGGCCTCAGGTATAAAG3’ and reverse primer 5’AAAGGGTTGATTGCGGAAAC3’)
cPCR-multiplex endogenous control: 431 bp of the HGH gene (forward primer 5’TGCCTTCCCAACCATTCCCTTA3’ and reverse primer 5’CCACTCACGGATTCTGTTGTGTTTC3’).
PCR was carried out in a final volume of 15 μl in a T100TM thermocycler (BioRad). PCR conditions were as follows: 0.7 μM for exon 5 and 0.2 μM for intron 5 and 7 SSP primers and 0.1 μM control primers, 1 × Coral Load PCR buffer (Qiagen, Hilden, Germany), 2.0–1.75 mM MgCl2 (only for exon 5 primers reaction, Qiagen, Hilden, Germany), 1.5% glycerol, 0.2 mM deoxyribonucleotide triphosphate (dNTP) (Invitrogen, São Paulo, Brazil), 0.5% Q Solution (only for intron 5 and 7 primers reaction, Qiagen, Hilden, Germany), 0.03 U/μl of Taq polymerase (Invitrogen, São Paulo, Brazil), 20 ng/μl DNA and water to complete the final volume. The amplification protocol starts with a 3 min denaturation step at 96°C, followed by 35 cycles of 15 sec at 94°C, 30 sec at the specific annealing temperature and 30 sec at 72°C, concluding with 5 min at 72°C in the final DNA extension step. Annealing temperature decreased every 10 cycles (64°C, 62°C and 60°C; 60°C, 58°C and 56°C for exon 5 and intron5_7 primers reaction, respectively), according to a previously published “touch-down” strategy which assures higher specificity to the amplification, while providing a larger amount of the desired PCR product [28]. The haplotypes defined by two SNPs, amplified by a pair of SSPs, were identified by the presence or absence of specific bands after agarose gel electrophoresis. Control bands informed on the quality of the reactions.
Ficolin-3 concentration assay
We measured ficolin-3 concentrations in 1:250 diluted sera (1:150 or 1:50 when necessary) of 61 patients and 73 controls with the same proportion of selected FCN3 genotypes, using the enzyme-linked immune sorbent assay HK 340 (Hycult Biotechnology, Uden, The Netherlands). Relative low ficolin-3 concentration was defined as <10368 ng/mL, which corresponded to the 20th percentile, and high levels as >33362 ng/mL, corresponding to the 80th percentile among controls.
Statistical analyses
Genotype and allele frequencies were obtained by direct counting. The hypothesis of Hardy–Weinberg equilibrium was verified using the approach of Guo and Thompson implemented in the ARLEQUIN software package version 3.1 (http://anthro.unige.ch/arlequin/). Tests of independence between patients and controls, as well as between patients with the lepromatous and non-lepromatous forms (tuberculoid, borderline and undetermined form of leprosy), were performed using Fisher exact test. Ficolin-3 levels were compared between the groups using nonparametric Mann-Whitney/Kruskal–Wallis tests using GraphPad Prism 3.0 software package. Two-tailed P-values less than 5% were considered significant. Logistic regression models were used to adjust results for age, sex and ethnic group distribution, using STATA v.9.2 (Statacorp, USA). Due to the sample size, statistical analyzes were performed between lepromatous leprosy and non-lepromatous patients (which included tuberculoid, borderline and undetermined leprosy). Clinical forms of leprosy was compared with healthy controls since this approach could reveal subtle differences not apparent when comparing them just with leprosy per se.
Results
FCN3 polymorphisms and haplotypes
FCN3 genotype distribution was in Hardy-Weinberg equilibrium. The allelic frequencies in Euro-Brazilian and Afro-Brazilian patients and controls did not differ from those reported in the HapMap project for CEU (North-Americans of Northern and Western European ancestry from Utah) and YRI (Yoruba in Ibadan, Nigeria) populations [29]. There was no difference in the allelic and genotypic frequencies between controls and leprosy patients, as well as lepromatous and non lepromatous groups (Table 2). Importantly, due to the very low frequencies of g.1637del, our study was underpowered in detecting associations with this SNP. Euro-Brazilians and Afro-Brazilians as well as males and females also had similar allelic and genotypic frequencies.
Table 2. FCN3 genotype, allele and haplotype frequencies (%).
| dbSNP | Controls | Leprosy per se | Lepromatous | Non-lepromatous | |
|---|---|---|---|---|---|
| rs532781899 | N = 245 | N = 190 | N = 144 | N = 53 | |
| Genotype | g.1637C/1637C | 96.0 | 95.0 | 95.0 | 94.0 |
| g.1637del/1637C | 4.0 | 5.0 | 5.0 | 6.0 | |
| n = 490 | n = 380 | n = 228 | n = 106 | ||
| Allele | g.1637C | 97.8 | 97.6 | 97.4 | 97.2 |
| g.1637del | 2.2 | 2.4 | 2.6 | 2.8 | |
| rs28362807 | N = 146 | N = 149 | N = 87 | N = 42 | |
| Genotype | g.3524_3532del/3524_3532del | 51.0 | 52.0 | 49.0 | 55.0 |
| g.3524_3532del/3524_3532ins | 42.0 | 42.0 | 43.0 | 43.0 | |
| g.3524_3532ins/3524_3532ins | 7.0 | 6.0 | 8.0 | 2.0 | |
| n = 292 | n = 298 | n = 174 | n = 84 | ||
| Allele | g.3524_3532del | 72.3 | 72.8 | 71 | 76.2 |
| g.3524_3532ins | 27.7 | 27.2 | 29 | 23.8 | |
| rs4494157 | N = 146 | N = 149 | N = 87 | N = 42 | |
| Genotype | g.4473C/4473C | 55.0 | 58.0 | 53.0 | 64.0 |
| g.4473C/4473A | 40.0 | 38.0 | 40.0 | 36.0 | |
| g.4473A/4473A | 4.0 | 5.0 | 7.0 | - | |
| n = 292 | n = 298 | n = 174 | n = 84 | ||
| Allele | g.4473C | 75.7 | 76.5 | 73 | 82.1 |
| g.4473A | 24.3 | 23.5 | 27 | 17.9 | |
| Phylogenetic nomenclature | Haplotype | n = 292 | n = 298 | n = 174 | n = 84 |
| *2ª | C del C | 71.6 | 71.1 | 69 | 73.8 |
| *2B1 | C ins A | 24.3 | 23.5 | 27 | 17.9 |
| *1 | C ins C | 2.74 | 3.7 | 2.3 | 5.9 |
| *2B2.2A | del del C | 0.68 | 1.7 | 1.7 | 2.4 |
| *2B2 | del ins C | 0.68 | - | - | - |
N: number of individuals. n: number of chromosomes. dbSNP: nomenclature according to the Single Nucleotide Polymorphism database. Unspecified leprosy patients were excluding when clinical forms of the disease were compared.
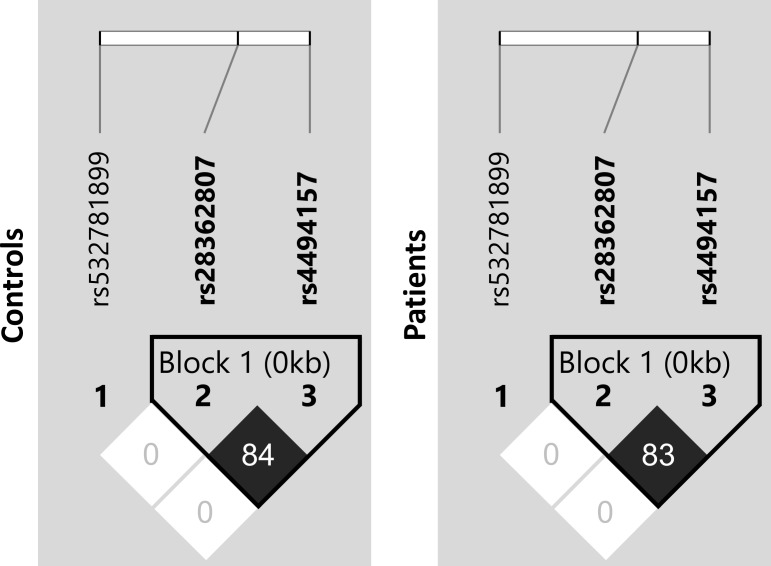
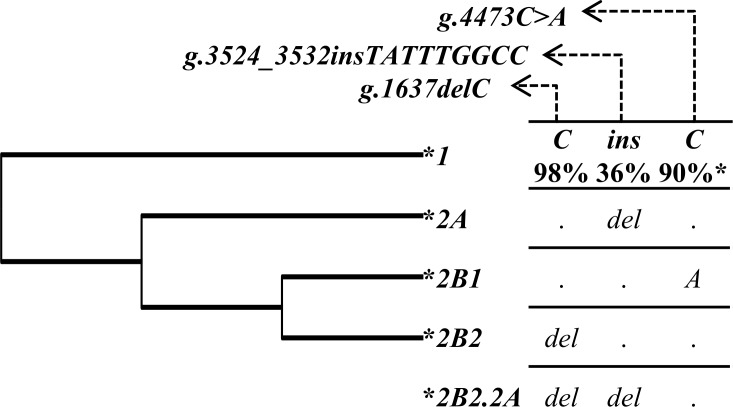
There was strong linkage disequilibrium (LD) between the two non-coding SNPs rs28362807 (g.3524_3532insTATTTGGCC, intron 5) and rs4494157 (g.4473C>A, intron 7), as indicated by the correlation coefficient values (r2) (Fig 1). The low r2 values observed for g.1637delC SNP reflect the completely discrepant frequencies of this SNP (an uncommon deletion) in comparison to the other two SNPs. Different combinations of investigated polymorphisms (g.1637delC, g.3524_3532insTATTTGGCC and g.4473C>A) resulted in 5 observed haplotypes, one of them possibly recombinant. According to the degree of sequence identity with the Pan troglodytes FCN3 gene sequence, the most probable ancestral haplotype (named as *1) is formed by g.1637C, g.3524_3532ins and g.4473C alleles (for short, CinsC) (Fig 2). The *2A (CdelC) haplotype was the most frequent, with 69–74% frequency in all investigated groups followed by *2B1 (CinsA) haplotype (18–27%). Other haplotypes, including *2B2 (delinsC), harboring the deletion in exon 5, were rather uncommon (Table 2). All the alleles found in our Afro-Brazilian sample were also found in all African population from NCBI and 1000 Genomes Project [14,29], giving us clues about the likely African origin of these polymorphisms.
Fig 1. Linkage disequilibrium between FCN3 single nucleotide polymorphisms.
LD was calculated based on the data for controls and leprosy patients. Black squares represent high LD and white low LD as measured by the correlation coefficient (r2) between sites, which values are shown inside of the squares. SNP identifiers are indicated on the abscissas.
Fig 2. Phylogenetic tree of FCN3 haplotypes.
The maximum parsimony tree was rooted on the haplotype shared with Pan troglodytes (ENSPTRT00000000796), named as *1, and the derived haplotype as *2, following the schema numerals/letters/ numerals, if they diverge further [30]. Recombinants are named according to the most common inferred parental haplotypes, separated by a dot [31]. SNPs in haplotypes were ordered according to their chromosomal position. *Allelic frequency in the African population, from the 1000 Genomes project [14].
Association of FCN3 polymorphisms and ficolin-3 levels
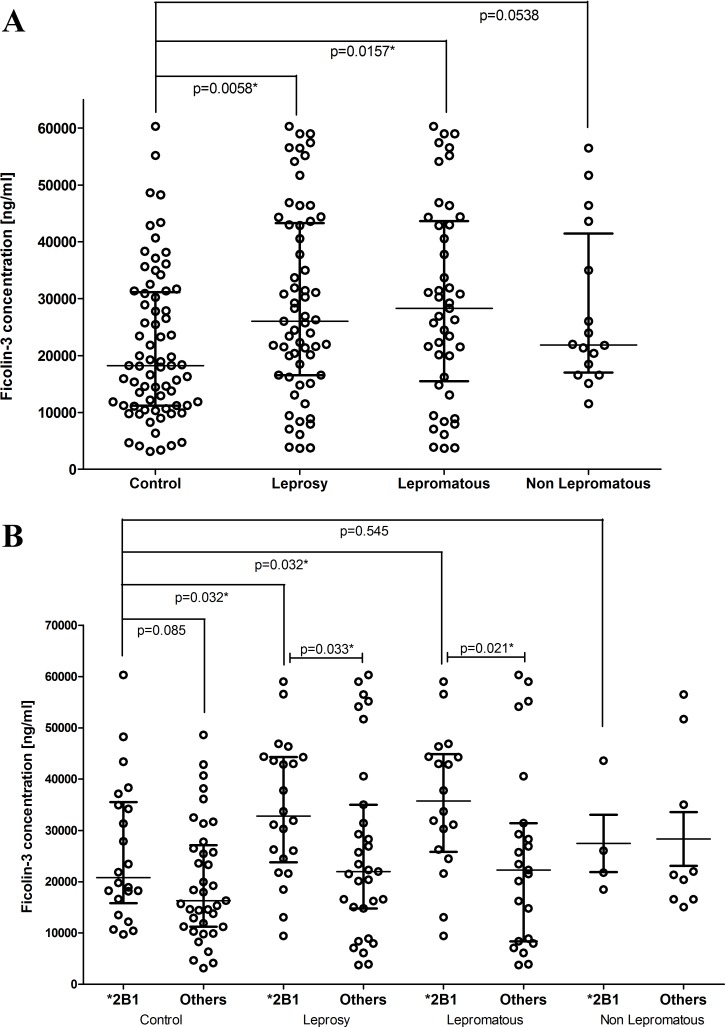
The median level of ficolin-3 observed in the control group (18231 ng/mL [3129–60300 ng/mL]) is in good agreement with published data in adults (~19500 ng/mL; [19]). The concentration of ficolin-3 in serum was higher in leprosy per se (26034 ng/mL, p = 0.005, OR 6.80 [1.65–28]) and lepromatous patients (28295 ng/mL, p = 0.016, OR 6.77 [1.43–32]) compared with controls (18231 ng/mL), even after correction for age, sex and ethnic group, but did not differ between lepromatous and non lepromatous groups (Fig 3A). In addition, high ficolin-3 levels (>33362 ng/mL) were more common in leprosy per se (34.4%) and in lepromatous patients (35.5%) than controls (19.2%; p = 0.045 and p = 0.047, respectively), while the frequency of low ficolin-3 concentrations (<10368 ng/mL) did not differ between the groups (Table 3). To assess whether the polymorphisms g.1637delC (rs532781899); g.3524_3532insTATTTGGCC (rs28362807) and g.4473C>A (rs4494157) of FCN3 gene were related to the variation of ficolin-3 serum concentration, they were evaluated separately and as haplotypes.
Fig 3. Distribution of Ficolin-3 levels in the investigated groups.
Ficolin-3 levels in (a) the investigated groups and (b) in individuals with and without the FCN3 *2B1 haplotype. Comparisons were made using Mann-Whitney test. * indicates significant p values (<0.05). Bars indicate median and interquartile values.
Table 3. Frequency of individuals with high and low ficolin-3 serum levels.
| Ficolin-3 Levels | Controls N = 73 | Leprosy per se N = 61 | Lepromatous N = 45 | Non lepromatous N = 16 | Controls vs. Leprosy per se | Controls vs. Lepromatous |
|---|---|---|---|---|---|---|
| High (%) | 19.2 | 34.4 | 35.5 | 31.2 | p = 0.0451 OR 2.21 [95% CI 1.01–4.85] |
p = 0.0471 OR 2.32 [95% CI 0.99–5.4] |
| Low (%) | 19.2 | 14.7 | 20.0 | - | n.s. | n.s. |
*Statistically significant value adjusted for age, sex and ethnic group. N: number of subjects. OR: Odds ratio. CI: confidence interval. n.s: non-significant. High ficolin-3 levels was defined as >33362 ng/mL, which corresponded to the 80th percentile, and low levels as <10368 ng/mL, corresponding to the 20th percentile among controls.
The g.1637del/1637C heterozygote controls had lower ficolin-3 median concentration (3762 ng/mL) than g.1637C/1637C homozygote controls (18382 ng/mL, p = 0.023), corroborating previous published data [17,32]. Surprisingly, we did not observe the same difference between leprosy patients (p = 0.143), moreover, in both g.1637del/1637C and g.1637C/1637C genotypes, patients had higher ficolin-3 concentration than controls (S1 Table). The effect of remaining polymorphisms in ficolin-3 levels was evaluated by removing all g.1637del/1637C individuals of the analyses. In the dominant model, ficolin-3 levels were higher in leprosy per se and lepromatous patients with g.4473A allele when compared to g.4473C/4473C (p = 0.043 and p = 0.028, respectively). Moreover, the g.3524_3532ins and g.4473A alleles were associated with higher ficolin-3 levels in leprosy patients, compared to controls (p = 0.042, p = 0.040; respectively). Under a recessive model, homozygous genotypes for rarer alleles (g.3524_3532ins and g.4473A) seem to lead to increased ficolin-3 in patients when compared to controls. All results were adjusted for demographic factors by logistic regression (S1 Table).
The same pattern of associations was observed in the haplotypes, with ficolin-3 levels being higher in leprosy patients with the *2B1 haplotype (CinsA), than in those without it (32795 vs. 21958 ng/mL, p = 0.033; excluding individuals with the deletion in exon 5), and than in *2B1 controls (20790 ng/ml; p = 0.032). Similarly, lepromatous patients with the *2B1 haplotype had higher ficolin-3 levels than lepromatous patients without it (35731 vs. 22294 ng/mL, p = 0.021) and than *2B1 controls (p = 0.032). There was a trend in the same direction, between controls with and without *2B1 (p = 0.080; Fig 3B).
Discussion
There are several evidences indicating an immunoregulatory role for the pattern recognition molecules (PRMs) of the lectin pathway in the susceptibility and clinical expression of leprosy [6,11,12,33]. This is the first study addressing FCN3 polymorphisms and ficolin-3 levels in leprosy. In previous studies, high MBL levels were shown to increase the susceptibility to lepromatous leprosy [6,34], whereas MBL2 haplotypes/genotypes conferring low MBL levels and deficiency in complement activation, conferred resistance against the development of lepromatous and borderline leprosy [33]. Furthermore, FCN2 and FCN1 haplotypes have protective effects against the susceptibility to leprosy per se [11,12]. Thus, components of the lectin pathway seem to be good candidates as biomarkers to be associated with the host response against M. leprae.
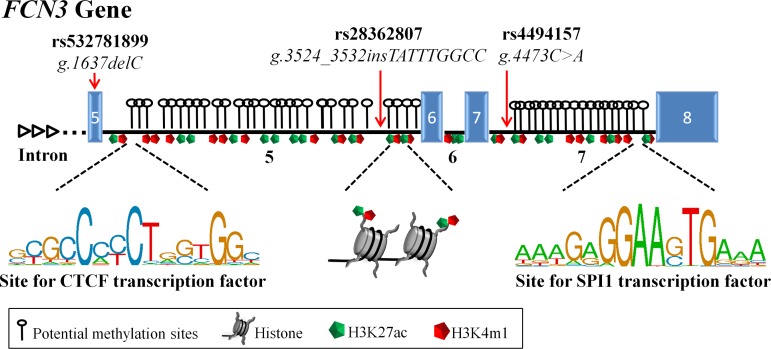
In this study, we observed that higher ficolin-3 levels were associated with the disease per se and with lepromatous leprosy. Higher ficolin-3 levels were also associated with a specific FCN3 haplotype, containing an insertion in intron 5 (g.3524_3532insTATTTGGCC) and the A allele at position +4473 in intron 7 (g.4473A). Interesting, introns 5 and 7 contain CpG islands. They are also enriched for typical histone modifications, known to characterize active enhancers (H3K27ac—H3 acetylated at lysine 27 and H3K4me1- H3 monomethylated at lysine 4) [35,36]. Different regulatory proteins (such as CTCF—CCCTC-binding factor, SPI1—Spleen focus forming virus (SFFV) Proviral Integration 1 and EGR1—Early growth response protein 1) bind to these intronic regions, as seen by chromatin immunoprecipitation assay in different cell lines (such as NHLF—lung fibroblasts, BJ—skin fibroblast and HMC—cardiac myocytes) [37]. Variants within these sequences, as those investigated here (or others strongly linked), may increase enhancer activity in response to inflammation signals, causing enhanced gene transcription and higher protein levels (Fig 4). This would explain why we only found clear evidence for an association in patients, which probably present an inflammatory response, observing only a trend in healthy individuals. On the other hand, we cannot dismiss the possibility that binding of regulatory proteins in these sites could modulate splicing of the alternative exon 4, whose inclusion in the most abundant FCN3 transcript leads to a longer collagenous tail.
Fig 4. Schematic representation of the regulatory intronic region of the FCN3 gene.
Introns 5 and 7 contain many potential methylation sites (CpG islands that may act as enhancers to transcription initiation), H3K27ac and H3K4me1 histone modifications (known to flank active enhancers) and sites to transcription factors known to regulate cell activation. CTCF—CCCTC-binding factor and SPI1—Spleen focus forming virus (SFFV) Proviral Integration 1, are the most evident regulatory proteins that may bind to intron 5 and 7, respectively, data from ENCODE [37]. SNPs investigated in the present work are indicated by the red arrows. The insertion allele in intron 5 (g.3524_3532insTATTTGGCC) and the A at position +4473 in intron 7 (g.4473A), may increase enhancer activity in response to inflammation signals, increasing gene transcription. Exons (blue boxes) and introns are drawn to scale.
High ficolin-3 levels have been previously reported in the sera of systemic lupus erythematosus patients [24], children with acute leukemia [21], ovarian cancer patients [25] and associated with graft loss in kidney transplant recipients [26], indicating a probable pathogenetic role for high ficolin-3 concentration in these disorders. We hypothesize that high ficolin-3 levels in leprosy patients probably play an unfavorable role by facilitating M. leprae dissemination. M. leprae may explore complement activation and opsonization induced by PRMs as one of the invasion mechanisms of macrophages and consequent evasion of the immune system [38]. Indeed, lectin pathway PRMs, including MBL (mannose-binding lectin) and ficolin-2, were shown to bind to mycobacteria leading to MASP2 (MBL-associated serine proteases) activation [38]. Although no direct binding of ficolin-3 on M. bovis BCG cell surface was found [38], it is know that the mycobacterial cell walls comprise long polymers of N-acetyl glucosamine (GlcNAc) [39], which is a ligand for ficolins [40], and could therefore be a potential target for ficolin-3.
The effect of the g.1637delC (rs532781899) polymorphism reducing ficolin-3 serum concentration [17] was only evident in controls. This is most probably a sampling effect, because among the two heterozygous g.1637del/1637C patients, one also carried the g.3524_3532ins and g.4473A alleles, associated with increased ficolin-3 concentration. Thus, whereas the FCN3 transcript with the g.1637del allele would produce a non-functional protein in this individual, the g.3524_3532ins and g.4473A alleles, combined in a haplotype harboring the wild type allele (g.1637C), form a functional protein and lead to high ficolin-3 concentration in this patient (21337 ng/mL), elevating the ficolin-3 mean level in the heterozygous g.1637del/1637C patients.
In conclusion, we identified high concentration of ficolin-3 in leprosy patients, associated with FCN3 polymorphisms present in introns 5 and 7. We suggest that high ficolin-3 levels increase the susceptibility to leprosy playing an unfavorable role in these patients by favoring M. leprae dissemination.
Supporting information
(DOCX)
The use of allele specific forward (g.3524_3532*del or g.3524_3532*ins, represented in dark and light blue arrows, respectively) and reverse (g.4473*A or g.4473*C, represented in light and dark green arrows, respectively) primers allow physical haplotype phasing. Each box represents one haplotype, specified in the upper left corner. DNA sequences are showed from 5' to 3', with one strand of the genomic DNA being represented, where bold letters represent the primer annealing region and polymorphisms are shown in red. For each tested sample, the four combinations of forward and reverse primers are tested in PCR. Amplification happens only if both primers anneal perfectly to the same chromosome. If only one of the four combination result in amplification, the individual is a homozygote for that specific haplotype. If instead two combinations result in amplification, the individual is heterozygote. Amplifications are visualized in an agarose gel after electrophoresis. Another, unrelated fragment is amplified in the same PCR as a control of PCR efficiency.
(TIF)
* Statistically significant value. aKruskal–Wallis tests. n.a. not applicable.
(DOCX)
Acknowledgments
We gratefully acknowledge the subjects of this investigation for their consent in participating in the study. We also thank the medical staff of the Hospital de Clínicas of the Federal University of Paraná and of the Sanitary and Dermatologic Hospital of Paraná for patient recruitment, and to the staff of the Laboratório de Imunopatologia Molecular in Curitiba for DNA extraction.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants and scholarships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Walker SL, Lockwood DNJ. Leprosy. Clin. Dermatol. 2007;25(2):165–72. 10.1016/j.clindermatol.2006.05.012 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Weekly epidemiological record Relevé épidémiologique hebdomadaire. 2013;88:365–380. [Google Scholar]
- 3.Misch EA, Berrington WR, Vary JC, Hawn TR. Leprosy and the human genome. Microbiol. Mol. Biol. Rev. 2010;74(4):589–620. 10.1128/MMBR.00025-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White C, Franco-Paredes C. Leprosy in the 21st century. Clin. Microbiol. Rev. 2015;28(1):80–94. 10.1128/CMR.00079-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mira MT. Genetic host resistance and susceptibility to leprosy. Microbes Infect. 2006;8(4):1124–1131. 10.1016/j.micinf.2005.10.024 [DOI] [PubMed] [Google Scholar]
- 6.Dornelles LN, Pereira-Ferrari L, Messias-Reason I. Mannan-binding lectin plasma levels in leprosy: deficiency confers protection against the lepromatous but not the tuberculoid forms. Clin. Exp. Immunol. 2006;145(3):463–8. 10.1111/j.1365-2249.2006.03161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krarup A, Sørensen UBS, Matsushita M, Jensenius JC, Thiel S. Effect of capsulation of opportunistic pathogenic bacteria on binding of the pattern recognition molecules Mannan-binding lectin, L-ficolin, and H-ficolin. Infect. Immun. 2005;73(2):1052–1060. 10.1128/IAI.73.2.1052-1060.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahagún-Ruiz A, Breda LCD, Castiblanco Valencia MM, Elias WP, Munthe-Fog L, Garred P, et al. Studies of the binding of ficolin-2 and ficolin-3 from the complement lectin pathway to Leptospira biflexa, Pasteurella pneumotropica and Diarrheagenic Escherichia coli. Immunobiology. 2015;220(10):1177–1185. 10.1016/j.imbio.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 9.Mason CP, Tarr AW. Human lectins and their roles in viral infections. Molecules. 2015;20(2):2229–71. 10.3390/molecules20022229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garred P, Honoré C, Ma YJ, Rørvig S, Cowland J, Borregaard N, et al. The genetics of ficolins. J. Innate Immun. 2010;2(1):3–16. 10.1159/000242419 [DOI] [PubMed] [Google Scholar]
- 11.De Messias‐Reason I, Kremsner PG, Kun JFJ. Functional Haplotypes That Produce Normal Ficolin‐2 Levels Protect against Clinical Leprosy. J. Infect. Dis. 2009;199(6):801–804. [DOI] [PubMed] [Google Scholar]
- 12.Boldt ABW, Sanchez MIN, Stahlke ERS, Steffensen R, Thiel S, Jensenius JC, et al. Susceptibility to leprosy is associated with M-ficolin polymorphisms. J. Clin. Immunol. 2013;33(1):210–9. 10.1007/s10875-012-9770-4 [DOI] [PubMed] [Google Scholar]
- 13.Genotype-Tissue Expression (GTEx), 2003–2012. Database: [Internt]. Accessed: http://www.gtexportal.org/home/gene/FCN3.
- 14.Ensembl genome browser. Database: [Internet]. Accessed: http://www.ensembl.org/Homo_sapiens/Gene/Variation_Gene/Table?db=core;g=ENSG00000142748;r=1:27369112-27374824.
- 15.Hummelshoj T, Fog LM, Madsen HO, Sim RB, Garred P. Comparative study of the human ficolins reveals unique features of Ficolin-3 (Hakata antigen). Mol. Immunol. 2008;45(6):1623–1632. 10.1016/j.molimm.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 16.Michalski M, Świerzko A St., Pągowska-Klimek I, Niemir ZI, Mazerant K, Domżalska-Popadiuk I, et al. Primary Ficolin-3 deficiency–Is it associated with increased susceptibility to infections? Immunobiology. 2015;220:711–713. 10.1016/j.imbio.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 17.Munthe-Fog L, Hummelshøj T, Ma YJ, Hansen BE, Koch C, Madsen HO, et al. Characterization of a polymorphism in the coding sequence of FCN3 resulting in a Ficolin-3 (Hakata antigen) deficiency state. Mol. Immunol. 2008;45(9):2660–2666. 10.1016/j.molimm.2007.12.012 [DOI] [PubMed] [Google Scholar]
- 18.Munthe-fog L, Sc M, Hummelshøj T, Ph D, Honoré C, Madsen HO, et al. Immunodeficiency Associated with FCN3 Mutation and Ficolin-3 Deficiency. 2009;2637–2644. [DOI] [PubMed]
- 19.Sallenbach S, Thiel S, Aebi C, Otth M, Bigler S, Jensenius JC, et al. Serum concentrations of lectin-pathway components in healthy neonates, children and adults: mannan-binding lectin (MBL), M-, L-, and H-ficolin, and MBL-associated serine protease-2 (MASP-2). Pediatr. Allergy Immunol. 2011;22(4):424–30. 10.1111/j.1399-3038.2010.01104.x [DOI] [PubMed] [Google Scholar]
- 20.Svendsen CB, Hummelshøj T, Munthe-Fog L, Milman N, Garred P, Laursen I a., et al. Ficolins and Mannose-Binding Lectin in Danish patients with sarcoidosis. Respir. Med. 2008;102(9):1237–1242. 10.1016/j.rmed.2008.04.012 [DOI] [PubMed] [Google Scholar]
- 21.Schlapbach LJ, Aebi C, Hansen AG, Hirt A, Jensenius JC, Ammann RA. H-ficolin serum concentration and susceptibility to fever and neutropenia in paediatric cancer patients. Clin. Exp. Immunol. 2009;157(1):83–9. 10.1111/j.1365-2249.2009.03957.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaffer T, Flogerzi B, Schoepfer AM, Seibold F, Müller S. Increased titers of anti-Saccharomyces cerevisiae antibodies in Crohn’s disease patients with reduced H-ficolin levels but normal MASP-2 activity. J. Crohns. Colitis. 2013;7(1):1–10. [DOI] [PubMed] [Google Scholar]
- 23.Prohászka Z, Munthe-Fog L, Ueland T, Gombos T, Yndestad A, Förhécz Z, et al. Association of Ficolin-3 with Severity and Outcome of Chronic Heart Failure. PLoS One. 2013;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen T, Munthe-Fog L, Garred P, Jacobsen S. Serum levels of ficolin-3 (Hakata antigen) in patients with systemic lupus erythematosus. J. Rheumatol. 2009;36(4):757–759. 10.3899/jrheum.080361 [DOI] [PubMed] [Google Scholar]
- 25.Szala A, Sawicki S, Swierzko AS, Szemraj J, Sniadecki M, Michalski M, et al. Ficolin-2 and ficolin-3 in women with malignant and benign ovarian tumours. Cancer Immunol. Immunother. 2013;62(8):1411–9. 10.1007/s00262-013-1445-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smedbråten Y V, Sagedal S, Mjøen G, Hartmann A, Fagerland MW, Rollag H, et al. High ficolin-3 level at the time of transplantation is an independent risk factor for graft loss in kidney transplant recipients. Transplantation. 2015;99(4):791–6. 10.1097/TP.0000000000000422 [DOI] [PubMed] [Google Scholar]
- 27.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int. J. Lepr. Other Mycobact. Dis. 1966;34(3):255–73. [PubMed] [Google Scholar]
- 28.Boldt ABW, Petzl-Erler ML. A new strategy for mannose-binding lectin gene haplotyping. Hum. Mutat. 2002;19(3):296–306. 10.1002/humu.10051 [DOI] [PubMed] [Google Scholar]
- 29.National Center for Biotechnology Information (NCBI). Database: [Internet]. Accessed: www.ncbi.nlm.nih.gov.
- 30.Nebert DW. Proposal for an Allele nomenclature system based on the evolutionary divergence of haplotypes. Hum. Mutat. 2002;20(6):463–472. 10.1002/humu.10143 [DOI] [PubMed] [Google Scholar]
- 31.Boldt ABW, Messias-Reason IJ, Meyer D, Schrago CG, Lang F, Lell B, et al. Phylogenetic nomenclature and evolution of mannose-binding lectin (MBL2) haplotypes. BMC Genet. 2010;11:38 10.1186/1471-2156-11-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalski M, Szala A, St. Swierzko A, Lukasiewicz J, Maciejewska A, Kilpatrick DC, et al. H-ficolin (ficolin-3) concentrations and FCN3 gene polymorphism in neonates. Immunobiology. 2012;217(7):730–737. 10.1016/j.imbio.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 33.De Messias-Reason IJ, Boldt ABW, Moraes Braga AC, Von Rosen Seeling Stahlke E, Dornelles L, Pereira-Ferrari L, et al. The association between mannan-binding lectin gene polymorphism and clinical leprosy: new insight into an old paradigm. J. Infect. Dis. 2007;196(9):1379–1385. 10.1086/521627 [DOI] [PubMed] [Google Scholar]
- 34.Garred P, Harboe M, Oettinger T, Koch C, Svejgaard A. Dual role of mannan-binding protein in infections: another case of heterosis? Eur. J. Immunogenet. 1994;21(2):125–31. [DOI] [PubMed] [Google Scholar]
- 35.Deaton A, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. 10.1101/gad.2037511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15(4):272–86. 10.1038/nrg3682 [DOI] [PubMed] [Google Scholar]
- 37.Encyclopedia of DNA Elements (ENCODE). Database [Internet]. Accessed: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr1%3A27697287-27699264&hgsid=502309425_rYKsJ2LDapXZnKsbIuMaheJfT09r.
- 38.Carroll M V., Lack N, Sim E, Krarup A, Sim RB. Multiple routes of complement activation by Mycobacterium bovis BCG. Mol. Immunol. 2009;46(16):3367–3378. 10.1016/j.molimm.2009.07.015 [DOI] [PubMed] [Google Scholar]
- 39.Kieser KJ, Rubin EJ. How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol. 2014;12(8):550–562. 10.1038/nrmicro3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsushita M. Ficolins: Complement-activating lectins involved in innate immunity. J. Innate Immun. 2009;2(1):24–32. 10.1159/000228160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
The use of allele specific forward (g.3524_3532*del or g.3524_3532*ins, represented in dark and light blue arrows, respectively) and reverse (g.4473*A or g.4473*C, represented in light and dark green arrows, respectively) primers allow physical haplotype phasing. Each box represents one haplotype, specified in the upper left corner. DNA sequences are showed from 5' to 3', with one strand of the genomic DNA being represented, where bold letters represent the primer annealing region and polymorphisms are shown in red. For each tested sample, the four combinations of forward and reverse primers are tested in PCR. Amplification happens only if both primers anneal perfectly to the same chromosome. If only one of the four combination result in amplification, the individual is a homozygote for that specific haplotype. If instead two combinations result in amplification, the individual is heterozygote. Amplifications are visualized in an agarose gel after electrophoresis. Another, unrelated fragment is amplified in the same PCR as a control of PCR efficiency.
(TIF)
* Statistically significant value. aKruskal–Wallis tests. n.a. not applicable.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.