Figure 3.
Additional Targets of Serine ADPr
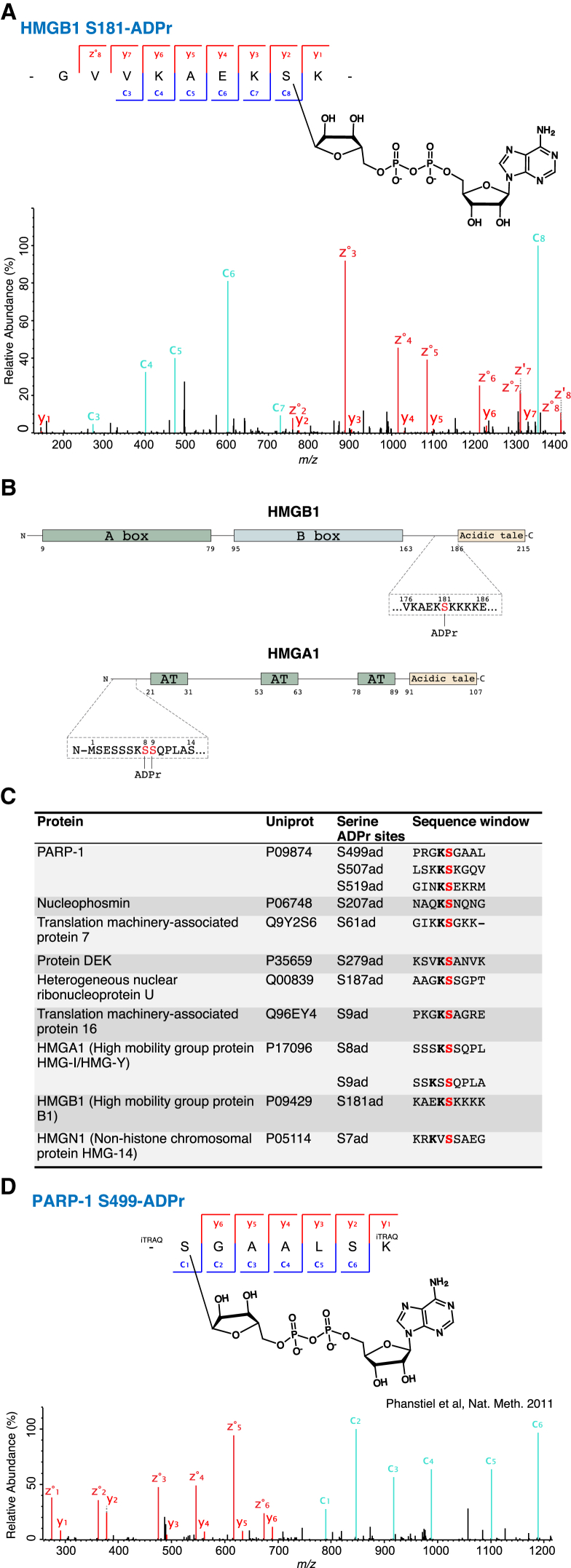
(A) High-resolution ETD fragmentation spectrum of an HMGB1 peptide with ADP-ribose on serine 181 is shown.
(B) Schematic representations of HMGB1 (upper) and HMGA1 (lower). The three novel serine ADPr sites were identified in vivo. A box and B box, positively charged homologous DNA-binding structures; acidic tail, negatively charged region composed of 30 glutamic and aspartic acids, exclusively; AT, AT-hook with the Arg-Gly-Arg-Pro (RGRP) core motif.
(C) Serine ADPr sites identified in histone-depleted fractions. ADPr sites were identified using ETD mass spectrometry. Modified serines are in red.
(D) High-resolution ETD fragmentation spectrum of a PARP-1 peptide with ADP-ribose on serine 499, obtained by reprocessing a published dataset (Phanstiel et al., 2011). The chemical structure of ADP-ribose is depicted.
See also Figure S3.