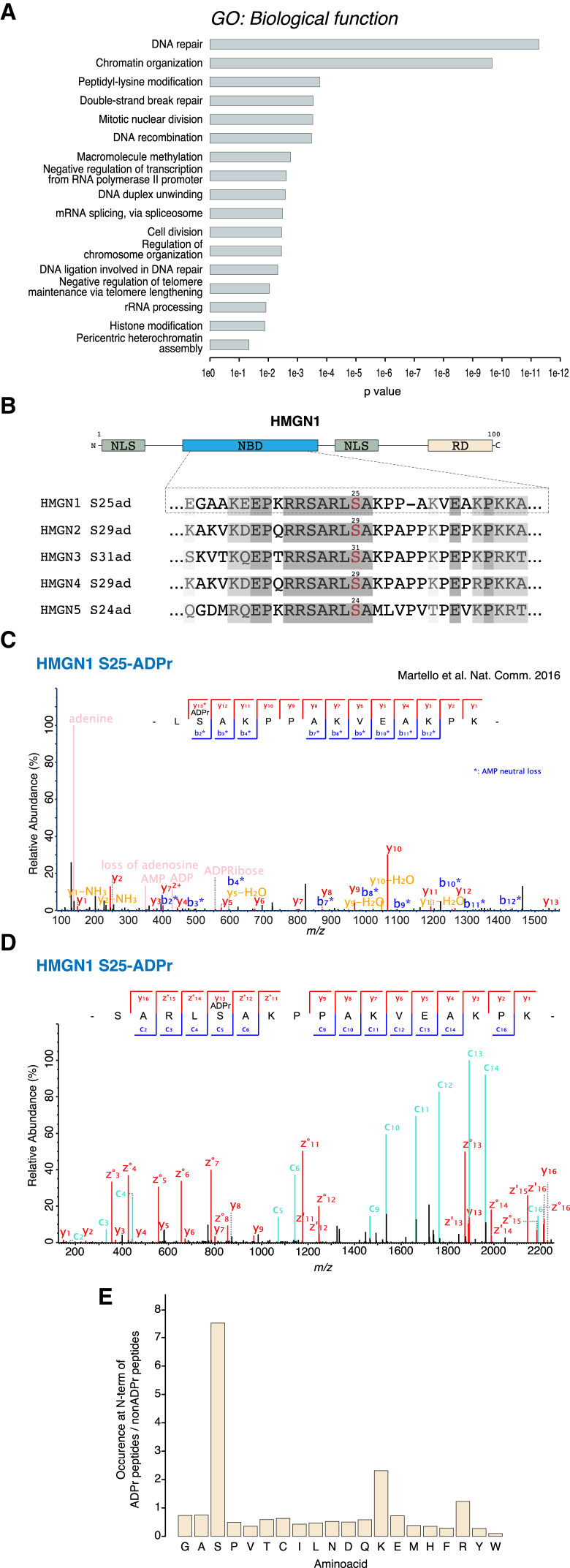
Figure 4.
Serine ADPr Is a Widespread PTM
(A) Gene ontology analysis of the serine ADPr proteins obtained by reprocessing a published dataset (Martello et al., 2016). Biological processes enriched in the serine ADPr proteins are shown.
(B) Schematic representation of HMGN1 (upper panel) and sequence alignment of the highly conserved N-terminal region of the HMGN family (lower panel). Note that the serine ADPr site (red) is conserved. NLS, nuclear localization signal; NBD, nucleosomal binding domain; RD, regulatory domain.
(C) HCD fragmentation spectrum of an HMGN1 peptide modified by ADP-ribose, obtained by reprocessing a published dataset (Martello et al., 2016). This HCD spectrum contains sufficient localization information to assign serine 25 as the modified residue. ∗AMP neutral loss.
(D) High-resolution ETD fragmentation spectrum of an HMGN1 peptide ADP-ribosylated on serine 25 in vitro in the presence of PARP-1 and HPF1 is shown.
(E) Bar plot shows the occurrence of the different amino acids at N termini of ADPr peptides relative to non-ADPr peptides, obtained by reprocessing a published dataset (Martello et al., 2016).
See also Figure S4.