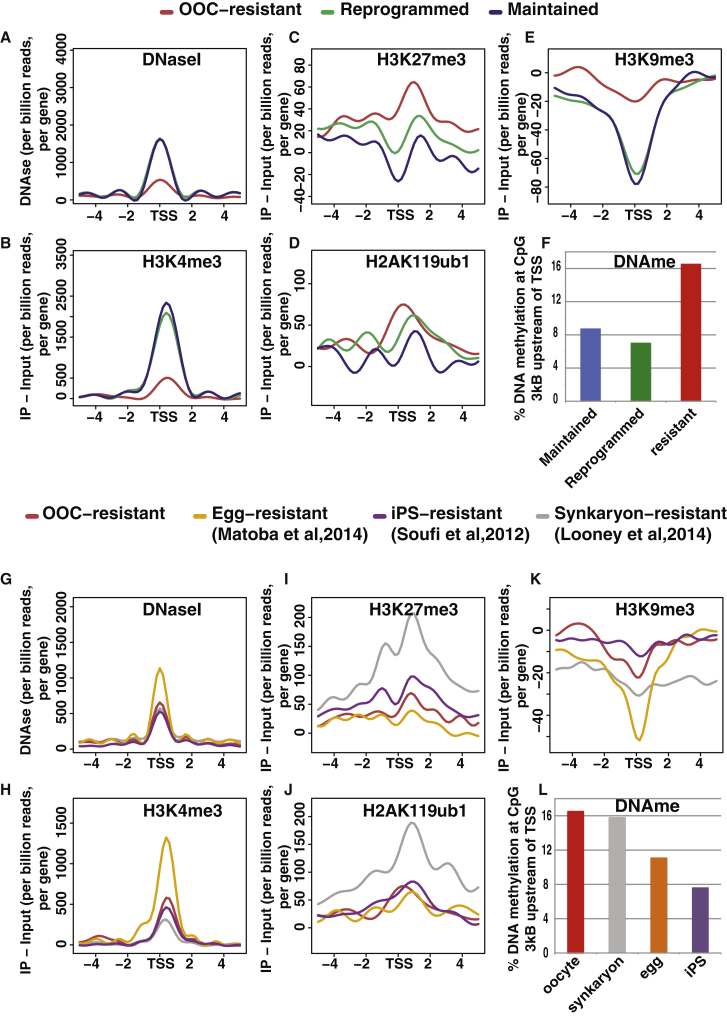
Figure 2.
Epigenetic Configuration of MEF Genes Resistant to OOC-Mediated Transcriptional Reprogramming
(A–E) Shown is the (A) DNase I sensitivity (Yue et al., 2014), (B) H3K4me3 (Kaneda et al., 2011), (C) H3K27me3 (Kaneda et al., 2011), (D) H2Aub (this study), and (E) H3K9me3 (Bulut-Karslioglu et al., 2014) metaplot analysis ±5 kB around the TSSs, in MEF, of genes with reprogrammed (green), maintained (blue), and resistant (red) expression following nuclear transfer.
(F) DNA methylation in the promoter region (3 kb upstream TSS) of reprogrammed, maintained, or resistant genes is shown as the percentage of CpG showing >95% methylation (data from Reddington et al., 2013).
(G–K) Shown is the (G) DNase I sensitivity (Yue et al., 2014), (H) H3K4me3 (Kaneda et al., 2011), (I) H3K27me3 (Kaneda et al., 2011), (J) H2Aub (this study), and (K) H3K9me3 (Bulut-Karslioglu et al., 2014) metaplot analysis ±5 kB around the TSSs of genes showing resistance in OOC-NT (red, from this study), mouse egg-NT (orange, from Matoba et al., 2014), transcription factor-induced reprogramming (purple, from Soufi et al., 2012), and cell fusion (gray, from Looney et al., 2014).
(L) DNA methylation in the promoter region (3 kb upstream TSS) of genes resistant in OOC-NT, mouse egg-NT, transcription factor-induced reprogramming, and cell fusion is shown as the percentage of CpG showing >95% methylation (data from Reddington et al., 2013). See also Figure S2.