Figure 1.
Mild Cooling Results in Translation Regulation of a Defined Set of Transcripts
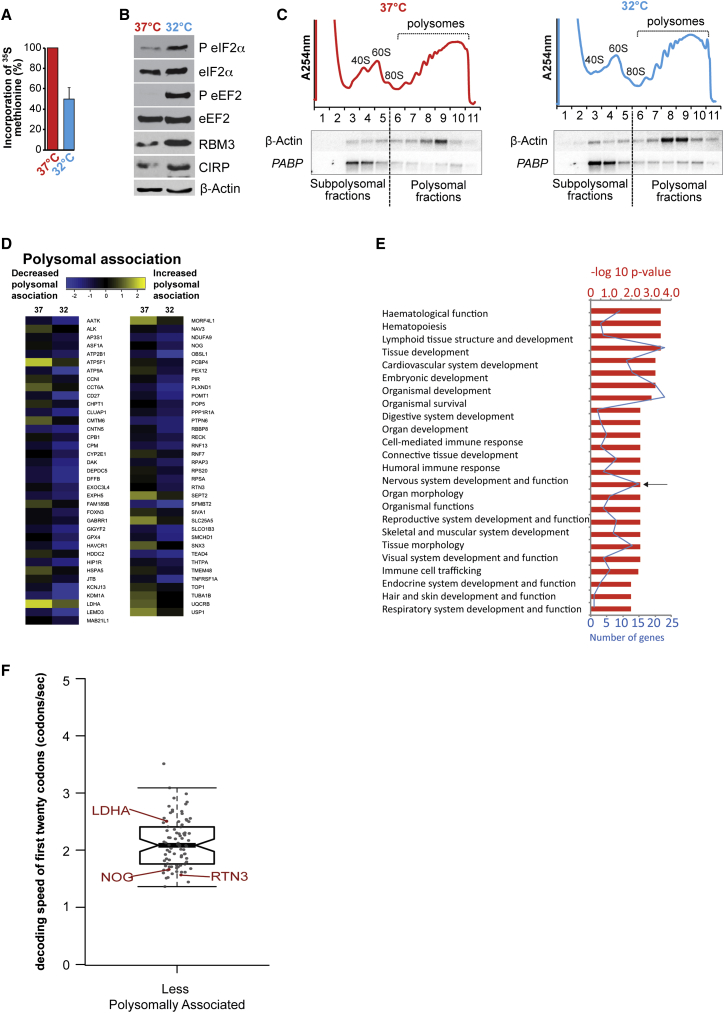
(A) Protein synthesis rates determined by [35S]-methionine label incorporation after 24 hr incubation of HEK293 cells at 32°C. Values were normalized to cells incubated at 37°C. Error bars represent SE within three independent experiments.
(B) HEK293 cells were incubated at 37°C or at 32°C for 24 hr and immunoblotted for RBM3 and CIRP, eIF2 alpha eEF2, and β-actin.
(C) Sucrose density gradient ultracentrifugations were performed from HEK293 cells incubated at 37°C or 32°C for 24 hr. Plots show the distribution of RNA within subpolysomes (40S, 60S, and 80S) and polysomes. Northern analysis was carried out on individual fractions, which were probed for β-actin or PABP.
(D) mRNAs from gradient fractions were pooled and subjected to cDNA microarray. The color scale represents the ratio of mRNA in subpolysome and polysome fractions, normalized log2 (polysome/subpolysome) value; yellow is polysome- and blue subpolysome-associated mRNAs.
(E) mRNAs that showed significant change in polysome/subpolysome (P/S) ratio on cooling were clustered into functional groups. Biological functions associated with decreased polysomal-associated transcripts, obtained from the ingenuity pathways analysis. Fisher’s exact test was used to calculate a p value (threshold p < 0.05) for each biological function represented in the red bar chart. The blue line represents number of proteins per category.
(F) Predictive modeling of transcript-decoding speed was performed on the initial 20 codons of human transcript sequences from those that showed decreased polysomal association. The boxplot shows mRNAs that have a decrease in polysomal association and contain an initial 20 “slow” codons (e.g., RTN3 and Noggin [NOG]) compared to those that contain “fast” codons, such as LDHA.
See also Figures S1–S3 and Tables S1–S6.