Figure 2.
RTN3 Is Subject to Elongation Control
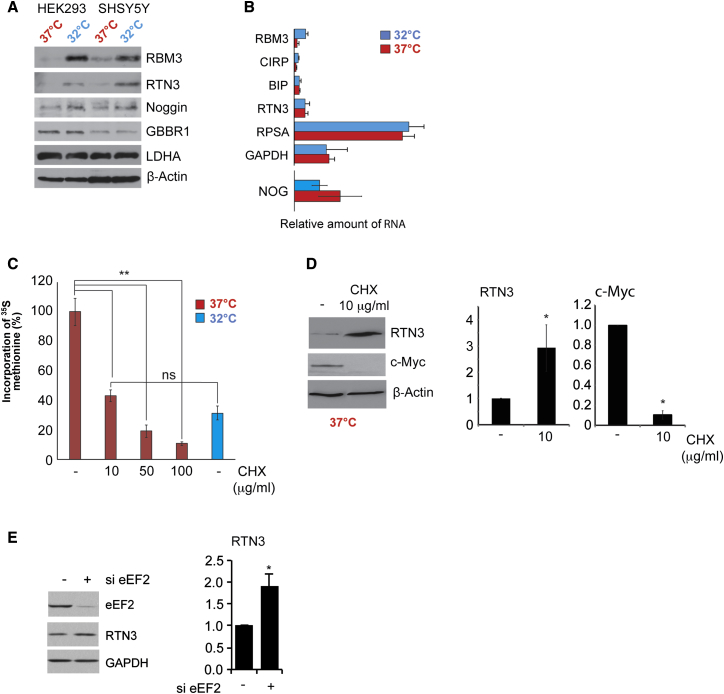
(A) Extracts from control or cooled HEK293 or SHSY5Y cells were immunoblotted for RTN3 and Noggin, GBBR1, and LDHA. β-actin is used as a loading control.
(B) qRT-PCR was performed on corresponding transcripts. Error bars represent 1 SD from the mean within three independent experiments. GAPDH was used as a control.
(C) Protein synthesis rates determined by [35S]-methionine label incorporation after 24 hr incubation of HEK293 cells at 37°C with cycloheximide. Values were normalized to untreated cells. A two-tailed paired Student’s t test was used to calculate statistical significance. Error bars represent SE within three independent experiments. ∗∗p < 0.001, all three conditions.
(D) Extracts from cells exposed to 10 μg/mL cycloheximide at 37°C were immunoblotted for RTN3, c-Myc, and β-actin. Error bars represent 1 SD from the mean within three independent experiments. ∗p < 0.01.
(E) eEF2 expression was reduced by siRNA, proteins extracted and immunoblotted with the antibodies shown. GAPDH was used as a loading control. Error bars represent 1 SD from the mean within three independent experiments. ∗p < 0.01.
See also Figures S4 and S5A.