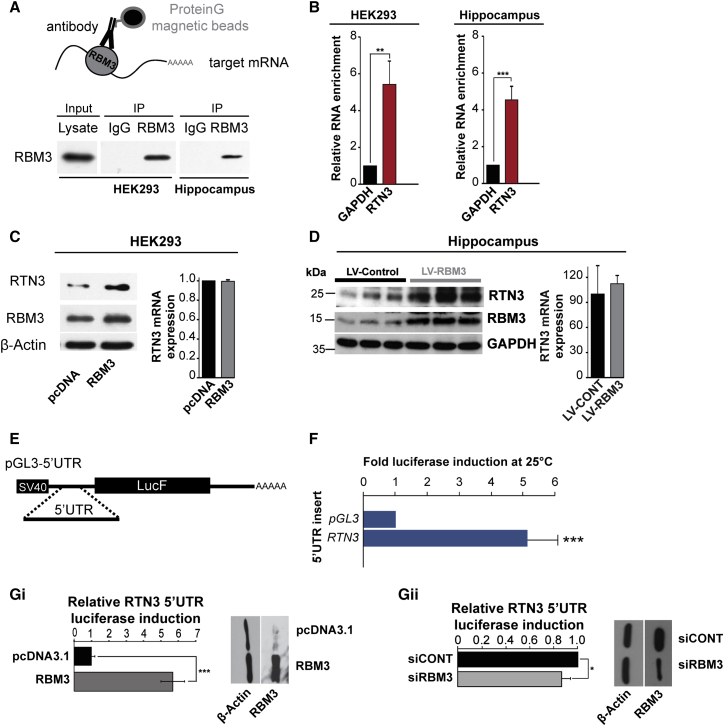
Figure 3.
RTN3 Expression Is Downstream of RBM3
(A) Schematic representation of an RNA immunoprecipitation (RNA-IP) assay. Immunoblots of input lysate from HEK293 cells or hippocampus, immunoprecipitated with either rabbit IgG or RBM3 antibody, are shown.
(B) qRT-PCR was performed on RNA-IP samples using primers specific for human (HEK293) or mouse (hippocampus) samples. All values are normalized with respect to the initial RNA input material, and the enrichment is plotted relative to GAPDH. A two-tailed paired Student’s t test was used to calculate statistical significance. Error bars represent 1 SD from the mean within three independent experiments. ∗∗p < 0.01; ∗∗∗p < 0.001.
(C) HEK293 cells were transfected with an expression plasmid construct encoding RBM3 or a control plasmid, and extracts were immunoblotted for RTN3. β-actin was used as a loading control. RNA expression of RTN3 was assessed by qRT-PCR.
(D) Mouse hippocampi stereotaxically injected with lentivirus containing a construct to overexpress RBM3 and extracts were immunoblotted for RTN3 and GAPDH. qRT-PCR was used to assess the expression of RTN3.
(E) Schematic representation of the RTN3 containing plasmid constructs encoding firefly luciferase.
(F) HEK293 cells were transfected with construct containing the 5′ UTR of RTN3 and a Renilla luciferase control and incubated at either 37°C or 25°C for 24 hr. Firefly luciferase activity was calculated relative to Renilla luciferase for each condition and expressed as the fold induction from 37°C to 25°C.
(Gi) HEK293 cells were transfected with either control (pcDNA3.1) or RBM3 expression plasmid (pcDNA-RBM3) and then transfected with either RTN3 5′ UTR pGL3 or pGL3 and Renilla luciferase constructs and luciferase activity determined. A two-tailed paired Student’s t test was used to calculate error. Error bars represent 1 SD from the mean within three independent experiments. ∗∗∗p < 0.001.
(Gii) HEK293 cells were transfected with either control siRNA (siCONT) or RBM3 siRNA (siRBM3) and then transfected with pGL3 and Renilla luciferase constructs. The fold repression from RTN3 5′ UTR pGL3 compared to the control pGL3 was calculated and normalized to siCONT transfection. A two-tailed paired Student’s t test was used to calculate statistical significance. Error bars represent 1 SD from the mean within three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.